Figures & data
Figure 2. Circular map of the complete mitochondrial genome of Thylacodes adamsii.
The complete mitochondrial genome is 14,913 bp in length and contains standard gene components. Of the 37 typical mitochondrial genes, 29 were located in the heavy strand (H-strand; genes named outside the circle) and the remaining 8 were in the light strand (L-strand; inside). The inner circle indicates the GC skew, which is the deviation from the average GC content of the entire mitogenome sequences.
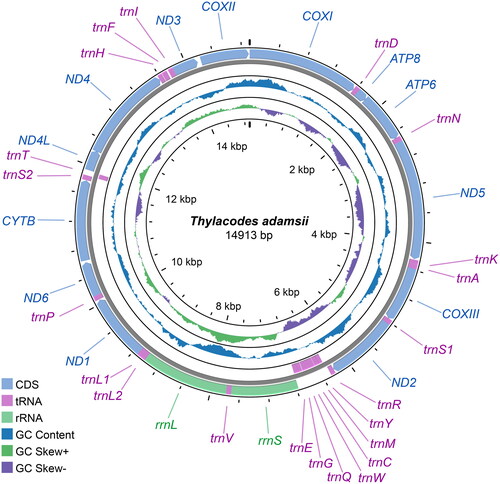
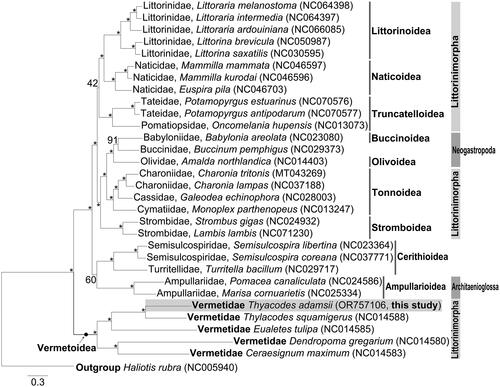
Figure 3. Maximum-likelihood tree inferred from nucleotide sequences of 13 PCGs of 30 caenogastropod species.
Maximum-likelihood tree showing the phylogenetic position of Vermetidae within Littorinimorpha. Haliotis rubra (Subclass Vetigastropoda) is used as an outgroup. The species examined in this study is presented in a gray box. The GenBank accession numbers of each species are indicated in parentheses. Numbers on the branches indicate nodal support bootstrapping values in percent. Branch supports are inferred from the ultrafast bootstrap method using IQ-TREE web server. The nodes exhibiting BP = 100 and mark an asterisk. The following sequences were used: Littoraria melanostoma NC064398 (Chen et al. Citation2023), Littoraria intermedia NC064397, Littoraria ardouiniana NC066085 (Wang et al. Citation2024), Littorina brevicula NC050987 (Bai et al. Citation2020), Littorina saxatilis NC030595 (Marques et al. Citation2017), Mammilla mammata NC046597 (Liu et al. Citation2020), Mammilla kurodai NC046596 (Liu et al. Citation2020), Euspira pila NC046703 (Liu et al. Citation2020), Potamopyrgus estuarinus NC070576 (Sharbrough et al. Citation2023), Potamopyrgus antipodarum NC070577 (Sharbrough et al. Citation2023), Oncomelania hupensis NC013073, Babylonia areolata NC023080 (Xiong et al. Citation2014), Buccinum pemphigus NC029373 (Xu et al. Citation2016), Amalda northlandica NC014403 (McComish et al. Citation2010), Charonia tritonis MT043269, Charonia lampas NC037188 (Cho et al. Citation2017), Galeodea echinophora NC028003 (Osca et al. Citation2015), Monoplex parthenopeus NC013247 (Cunha et al. Citation2009), Strombus gigas NC024932 (Márquez et al. Citation2014), Lambis lambis NC071230 (Li et al. Citation2022), Semisulcospira libertina NC023364 (Zeng et al. Citation2014), Semisulcospira coreana NC037771 (Kim and Lee Citation2018), Turritella bacillum NC029717 (Zeng et al. Citation2016), Pomacea canaliculata NC024586 (Zhou et al. Citation2016), Marisa cornuarietis NC025334 (Wang and Qiu Citation2016), thylacodes squamigerus NC014588 (Rawlings et al. Citation2010), Eualetes tulipa NC014585 (Rawlings et al. Citation2010), Dendropoma gregarium NC014580 (Rawlings et al. Citation2010), Ceraesignum maximum NC014583 (Rawlings et al. Citation2010), Haliotis rubra NC005940 (Maynard et al. Citation2005), and Thylacodes adamsii OR757106.
Supplemental Material
Download PDF (453.8 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession number OR757106. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1070832, SRR27775017, and SAMN39662919, respectively.

