Figures & data
Figure 1. Image of Perforatus perforatus collected from Mulchi-port, Yangyanggun, Gangwondo Province, Korea (38°9’20” N, 128°36’35” E). The photo was taken by Hyun Kyong Kim with a digital camera (model 5D mark IV; canon, Tokyo, Japan), and produced with helicon focus software (scale bar: 5 mm).

Figure 2. Mitochondrial genome map of perforatus perforatus obtained by the GenomeVx tool. tRNAs are abbreviated by the one-letter code of the corresponding amino acid with the anticodon within the parenthesis. Gene names located on the outside of the large circle are encoded on the heavy strand (5’-3’ direction) excluding control region, while gene names located inside are encoded on the light strand (3’-5’ direction). Color codes: green for PCGs, blue for tRNAs, orange for rRNAs, and white for control region.
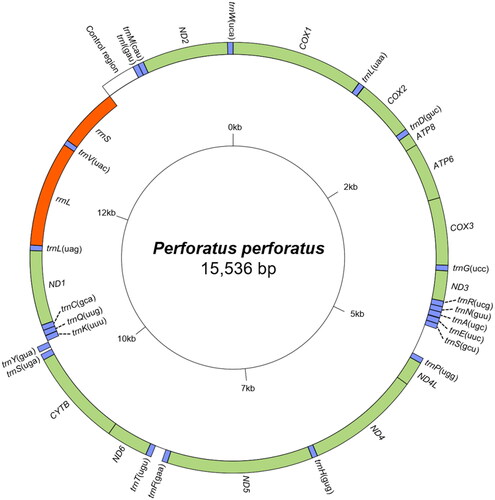
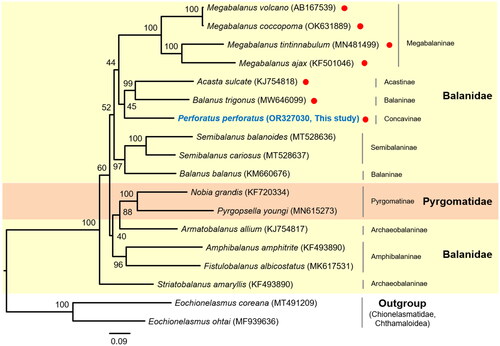
Figure 3. The phylogenetic tree of two families in balanoidea (Balanidae and pyrgomatidae) within balanoidea. The tree was obtained using concatenated nucleotide sequences of 13 protein-coding genes and two rRNAs using the maximum likelihood method. The numbers at each node indicate the bootstrap values. The scale bar indicates the number of substitutions per site. Two chionelasmatidae species (Eochionelasmus coreana and E. ohtai) in chthamaloidea were used as outgroups. Red dots indicate the species with an inverted sequence block (trnP-ND4L-ND4-trnH-ND5-trnF) between trnS(gct) and trnT. The following sequences were used: Acasta sulcate KJ754818 (Unpublished); Balanus trigonus MW646099 (Liu et al. Citation2021); Perforatus perforatus OR327030 (This study); Megabalanus tintinnabulum MN481499 (Feng et al. Citation2019); Megabalanus ajax KF501046 (Shen et al. Citation2016); Megabalanus volcano AB167539 (Unpublished); Megabalanus coccopoma OK631889 (Zhang et al. Citation2023); Semibalanus cariosus MT528637 (Nunez et al. Citation2021); Semibalanus balanoides MT528636 (Nunez et al. Citation2021); Balanus balanus KM660676 (Shen et al. Citation2016); Pyrgopsella youngi MN615273 (Unpublished); Nobia grandis KF720334 (Shen et al. Citation2016); Armatobalanus allium KJ754817 (Unpublished); Amphibalanus amphitrite KF493890 (Shen et al. Citation2015); Fistulobalanus albicostatus MK617531 (Song et al. Citation2020); Striatobalanus amaryllis KF493890 (Tsang et al. Citation2015); Eochionelasmus coreana MT491209 (Lee et al. Citation2021); and Eochionelasmus ohtai MF939636 (Kim et al. Citation2017).
Supplemental Material
Download PDF (668.5 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov, under the accession number OR327030. The associated BioProject, SRA, and Bio-Sample numbers: PRJNA1057449, SRR27587530, and SAMN39129815, respectively.
