Figures & data
Figure 1. Isolation of Mucor indicus mycelia from a wine fermentation system. A photo of the species was taken by Qiang Li.
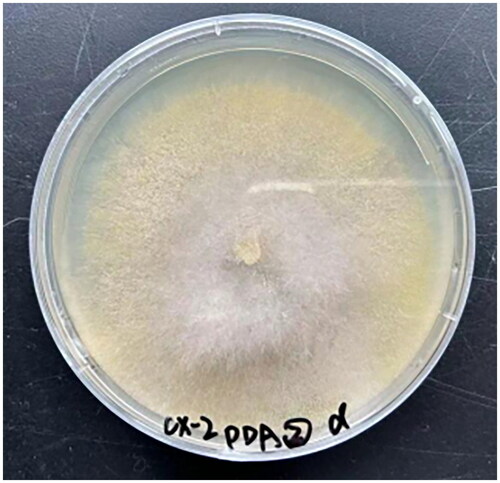
Figure 2. The circular mitochondrial genome map of Mucor indicus. Colored blocks outside each ring indicate that the genes are on the direct strand, while colored blocks within the ring indicate that the genes are located on the reverse strand. The inner grayscale bar graph shows the GC content of the mitochondrial sequences. The circle inside the GC content graph marks the 50% threshold.
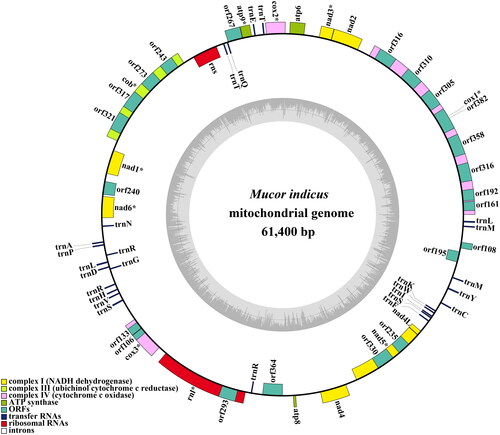
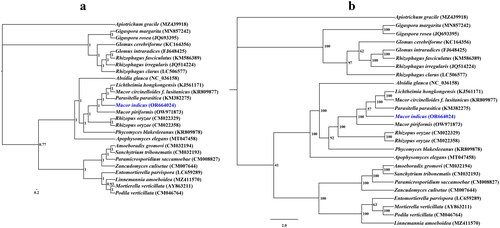
Figure 3. Bayesian inference (a) and maximum likelihood (b) tree generated using 14 concatenated mitochondrial protein-coding genes (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) from Mucor indicus and 25 other fungal species. The support values are Bayesian posterior probabilities (BPPs) and bootstrap values (BSs). Apiotrichum gracile was used as the outgroup. The accession number information of the sequence is as follows: Linnemannia amoeboidea (MZ411570) (Yang et al. Citation2022), Glomus cerebriforme (KC164356) (Beaudet et al. Citation2013), Mucor piriformis (OW971873), Rhizophagus fasciculatus (KM586389), Rhizophagus clarus (LC506577) (Kobayashi et al. Citation2018), Lichtheimia hongkongensis (KJ561171) (Leung et al. Citation2014), Gigaspora rosea (JQ693395) (Nadimi et al. Citation2012), Sanchytrium tribonematis (CM032193) (Galindo et al. Citation2021), Apiotrichum gracile (MZ439918) (Li et al. Citation2023), Mucor indicus (OR664024), Glomus intraradices (FJ648425) (Lee and Young Citation2009), Phycomyces blakesleeanus (KR809878), Entomortierella parvispora (LC659289) (Herlambang et al. Citation2022), Mortierella verticillata (AY863211) (Seif et al. Citation2005), Podila verticillata (CM046764) (Morales et al. Citation2022), Absidia glauca (NC_036158) (Ellenberger et al. Citation2016), Apophysomyces elegans (MT047458), Rhizopus oryzae (CM022358), Zancudomyces culisetae (CM007644) (Nie et al. Citation2019), Parasitella parasitica (KM382275) (Ellenberger et al. Citation2014), Paramicrosporidium saccamoebae (CM008827) (Quandt et al. Citation2017), Mucor circinelloides f. lusitanicus (KR809877), Amoeboradix gromovi (CM032194) (Galindo et al. Citation2021), Rhizophagus irregularis (JQ514224) (Formey et al. Citation2012), Rhizopus oryzae (CM022329), and Gigaspora margarita (MN857242) (Venice et al. Citation2020).
Supplemental Material
Download MS Word (486.9 KB)Supplemental Material
Download PDF (953.7 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov/under accession no. OR664024. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1025866, SRR26320847 and SAMN37723662, respectively.
