Figures & data
Figure 1. Photo of S. buxifolium. Lichai Yuan took the picture at Zhaiqian town, Guidong County, Chenzhou City, Hunan Province, China (N25˚59′26ʺ, E113˚54′39ʺ). A: Leaf; B: Inflorescence; C: branchlet.

Figure 2. The genomic map of the chloroplasts in S. buxifolium. There are six tracks on the map. The first track (A) displays the forward and backward repeats connected by red and green arcs, viewed from the center outward. The tandem repeats are displayed as brief blue bars on track B, the second track. The microsatellite sequences are displayed as brief green and yellow bars on the third track (C). On the fifth track (D), the small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) areas are displayed. Plotting of the GC content across the genome is done on track six (E). Plotting of genes is done on track seven (F). The gene name is followed by parenthesis with the optional codon use bias. Depending on how they are classified functionally, genes are colored. Transcription proceeds clockwise and counterclockwise for the inner and outer genes, respectively. The lower left corner displays the genes’ functional categorization.
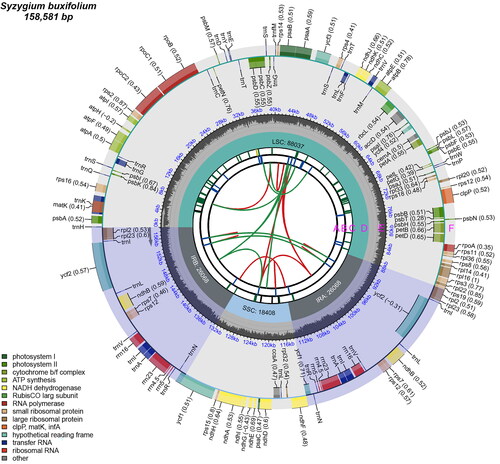
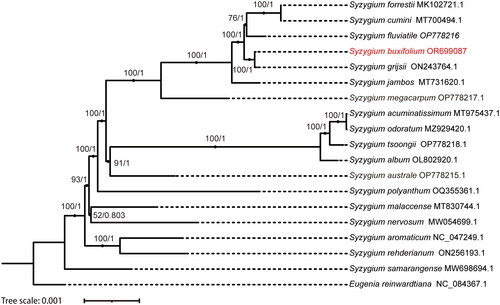
Figure 3. Phylogenetic tree generated using maximum likelihood (ML) Bayesian interference (BI) and based on the chloroplast genome. ML bootstrap (BS) and BI posterior probability (PP) values are represented by numbers at the nodes. The 17 Syzyglium were downloaded from GenBank, namely S. forrestii (MK102721.1) (Zhang et al. Citation2018), S. cumini (MT700494.1), S. fluviatile (OP778216.1), S. buxifolium (OR699087), S. grijsii (ON243764.1), S. jambos (L.) Alston (MT731620.1) (Sun et al. Citation2020), S. megacarpum Rathakr. & N. C. Nair (OP778217.1), S. aromaticum (L.) Merr. & L.M.Perry (NC_047249.1), S. rehderianum Merr. & L. M. Perry (ON256193.1), S. samarangense (Blume) Merr. & L. M. Perry (MW698694.1) (Wei et al. Citation2022), S. nervosum DC. (MW054699.1) (Li et al. Citation2021), S. malaccense (L.) Merr. & L. M. Perry (MT830744.1), S. polyanthum (Wight) Walp. (OQ355361.1), S. australe (J. C. Wendl. ex Link) B. Hyland (OP778215.1), S. acuminatissimum (Blume) DC. (MT975437.1) (Zeng et al. Citation2021), S. odoratum (Lour.) DC. (MZ929420.1) (Chen et al. Citation2022), S. tsoongii (Merr.) Merr. & L. M. Perry (OP778218.1), S. album Q. F. Zheng (OL802920.1), Eugenia reinwardtiana (NC_084367.1).
Data availability statement
The complete chloroplast genome sequences of S. buxifolium in this study have been submitted to the NCBI database under the accession number OR699087. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA1055315, SAMN38989340, and SRS19952658.
