Figures & data
Figure 1. Panorama (A) and detail (B) photos of Amaranthus roxburghianus.
Photo by Liqiang Wang, plant coordinates 35°16′23.65'' N, 115°27′32.85'' E. Main identifying features: stem erect, light green, 30-65 cm, much branched, glabrous. Petiole 1-2.5 cm, slender; leaf blade ovate-rhombic, obovate, or oblong, 2-5 × 1-2.5 cm, base cuneate, margin undulate, apex notched, mucronate. Flowers few, sparsely clustered in axils. Bracts and bracteoles subulate, ca. 2 mm, abaxially with distinct midvein, apex long pointed. Tepals lanceolate, ca. 2.5 mm, apex acuminate, long pointed. Stamens shorter than perianth; stigmas 3. Utricles ovoid, subequal to perianth, ca. 3 mm, circumscissile. Seeds brownish black, subglobose, ca. 1 mm in diameter. The scale of the scale bar is 10 cm.

Figure 2. The schematic map of the Amaranthus roxburghianus plastome. The species name is shown at the top left of the map. The map displays six tracks representing various genetic elements. Track one illustrates dispersed repeats, with direct and palindromic repeats connected by red and green arcs. Long tandem repeats in track two are shown as short blue bars. Track three depicts short tandem repeats/microsatellite sequences, differentiated by colored bars: black for complex repeats and green, yellow, purple, blue, orange, and red for repeat units of sizes 1 to 6, respectively. The fourth track indicates single-copy and inverted repeat regions, while the fifth track presents the genome’s GC content. The sixth track details genes, color-coded by functional classification and potentially including codon usage bias, with gene transcription direction indicated (inner genes clockwise, outer genes anticlockwise). The functional classification of genes is provided in the bottom left corner.
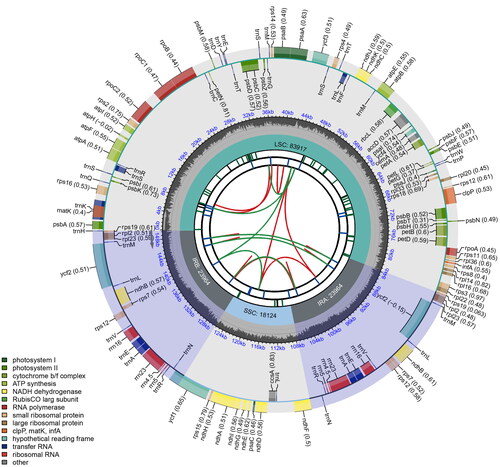
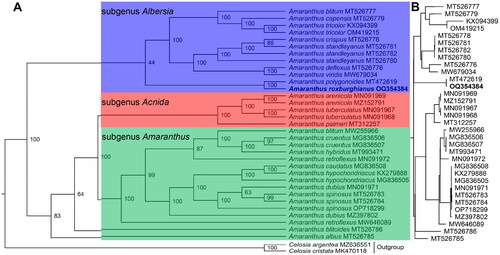
Figure 3. The maximum likelihood phylogeny of Amaranthus roxburghianus and its close relatives using whole plastome sequences. The bootstrap values based on 1000 replicates were shown on each node in the cladogram tree (a). The corresponding phylogram tree is shown in panel B without outgroups. The tree was constructed with 35 species, they were A. albus (MT526785) (Xu et al. Citation2022), A. arenicola (MN091969) (Xu et al. Citation2022), A. arenicola (MZ152791) (Xu et al. Citation2022), A. blitoides (MT526786) (Xu et al. Citation2022), A. blitum (MT526777) (Xu et al. Citation2021), A. blitum (MW255966), A. capensis (MT526779) (Xu et al. Citation2021), A. caudatus (MG836508), A. cruentus (MG836506), A. cruentus (MG836507), A. deflexus (MT526776) (Xu et al. Citation2021), A. dubius (MN091971) (Xu et al. Citation2022), A. dubius (MZ397802) (Xu et al. Citation2021), A. hybridus (MT993471) (Bai et al. Citation2021), A. hypochondriacus (KX279888), A. hypochondriacus (MG836505) (Xu et al. Citation2022), A. palmeri (MT312257) (Xu et al. Citation2022), A. polygonoides (MT472619) (Xu et al. Citation2022), A. retroflexus (MN091972) (Xu et al. Citation2022), A. retroflexus (MW646089) (Lou and Fan Citation2021), A. roxburghianus (OQ354384, this study), A. spinosus (MT526783) (Xu et al. Citation2022), A. spinosus (MT526784) (Xu et al. Citation2022), A. spinosus (OP718299) (Xu et al. Citation2022), A. crispus (MT526778) (Xu et al. Citation2021), A. standleyanus (MT526780) (Xu et al. Citation2021), A. standleyanus (MT526781) (Xu et al. Citation2021), A. standleyanus (MT526782) (Xu et al. Citation2021), A. tricolor (KX094399) (Xu et al. Citation2022), A. tricolor (OM419215) (Xu et al. Citation2022), A. tuberculatus (MN091967) (Xu et al. Citation2022), A. tuberculatus (MN091968) (Xu et al. Citation2022), A. viridis (MW679034) (Ding et al. Citation2021), Celosia cristata (MK470118, outgroup), and C. argentea (MZ636551, outgroup). Bootstrap supports were calculated from 1000 replicates. The A. roxburghianus was labeled in bold font in the phylogenetic tree. The blue, red and green box represents subgenus Albersia, subgenus Acnida and subgenus Amaranthus in the Amaranthus.
Supplemental Material
Download MS Word (4 MB)Data availability statement
The plastome sequence has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession number of OQ354384 (https://www.ncbi.nlm.nih.gov/nuccore/OQ354384). The associated BioProject, Bio-Sample and SRA numbers are PRJNA928567, SAMN36852648 and SRR23251385. (https://www.ncbi.nlm.nih.gov/sra/?term=SRR12620715).
