Figures & data
Figure 1. The morphological characteristics of P. major in our research. It is a perennial herb with basal leaves. Photoed by Yingying Liu in Guangxi Botanical Garden of Medicinal Plants, Nanning city, China.

Figure 2. Circular map of the complete chloroplast genome of P. major generated by CPGview. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
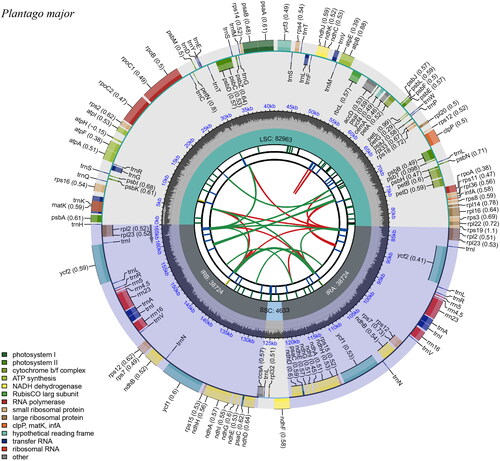
Figure 3. Maximum-likelihood phylogenetic tree of 25 plantago species was constructed based on 76 CDS sequences from chloroplast genomes of P. major and its closely related specie, with Crepidiastrum sonchifolium and Saxifraga perpusilla as outgroups. The number on each branch indicates the boot support value. The following sequences were used: P. asiatica NC 060521 (Si et al. Citation2022), P. fengdouensis NC 047190 (Wang et al. Citation2020), P. depressa NC 041161 (Kwon et al. Citation2019), P. rugelii MW 877573, P. australis MW 877569, P. rigida MW 877572, P. elongata MW 877571, P. tenuiflora MW 877574, P. coronopus MW 877566, P. macrorhiza MW 877568, P. crassifolia MW 877567, P. alpina MW 877565, P. atrata MW77580, P. amplexicaulis MW 877576, P. indica MW 877577, P. afra MW 877575, P. brasiliensis MW 877579, P. nubicola MW 877564, P. sericea MW 877584 (Mower et al. Citation2021), P. media NC 028520, P. maritima NC 028519 (Zhu et al. Citation2016), P. lagopus NC 041420, P. ovata NC 041421 (Sun et al. Citation2019), P. lanceolata NC 068049 (Zhao et al. Citation2023), C. sonchifolium NC 046513 (Cho et al. Citation2020), and S. perpusilla NC 070499 (Yuan et al. Citation2023).
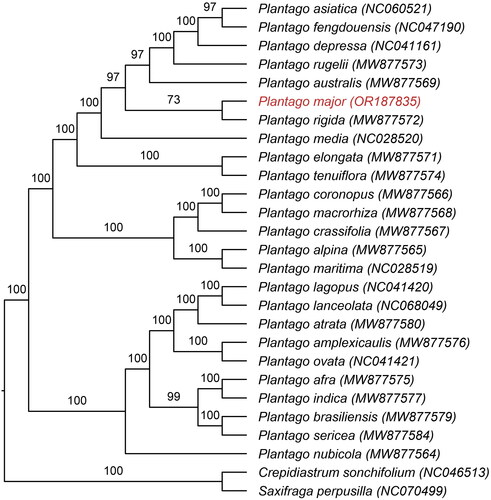
Figure 3.jpg
Download JPEG Image (1.5 MB)Figure 2.jpg
Download JPEG Image (2 MB)Figure 1.jpg
Download JPEG Image (4.2 MB)Manuscript clean.docx
Download MS Word (755.2 KB)Supplementary figures.docx
Download MS Word (196.2 KB)Data availability statement
The data of this study are openly accessible in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession number OR187835. The associated BioProject, SRA, and BioSample numbers are PRJNA987869, SRR25033257, and SAMN35994512, respectively.
