Figures & data
Figure 1. Bayesian chloroplast tree inferred with MrBayes. Posterior probabilities are indicated near nodes when greater than 0.8. The major clades within Themeda are delimited on the right (A–H). Within T. triandra, branches are coloured based on geographical origin. Scale bar shows the expected number of substitutions per site.
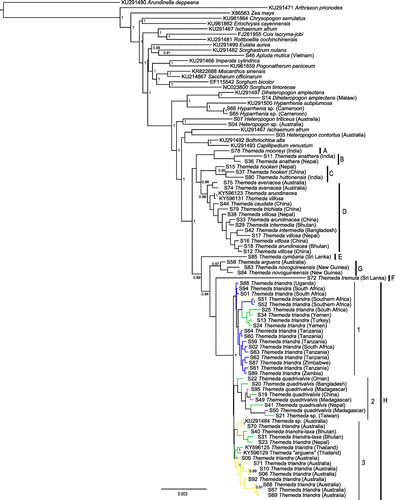
Table 1. Themeda species used in this study and their distribution. In total, we sequenced 18 out of the 27 known species. The clade assignment is based on chloroplast and nuclear markers, with the number of samples sequenced indicated (n), and ecological information derived from Morales (Citation2014).
Figure 2. Dating the divergence within Themeda using chloroplast data. Posterior probabilities are indicated near nodes (>0.8), and bars represent the 95% HPD of the estimated dates. The clade that each Themeda species represents is indicated (A–H; Table ). NCBI GenBank accession numbers are given for samples sequenced in previous studies. Within T. triandra, branches are coloured based on geographical origin. The scale is given at the bottom for a scenario that incorporates or ignores the microfossils. The origins of C4 grasslands are plotted based on each scenario.
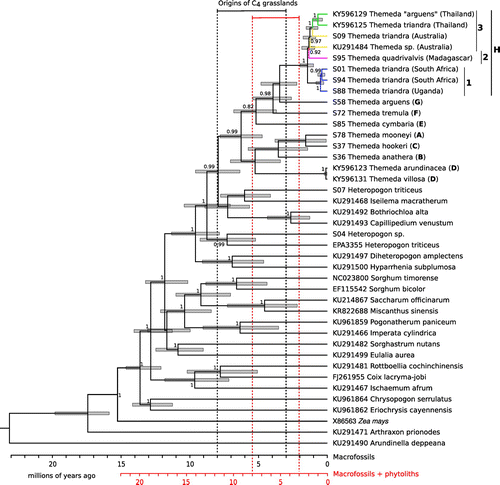
Figure 3. Nuclear genome relationships within Themeda. A Bayesian tree was inferred from genome-wide SNPs with MrBayes. Posterior probabilities are indicated near nodes. Branches are coloured based on geographical origin.
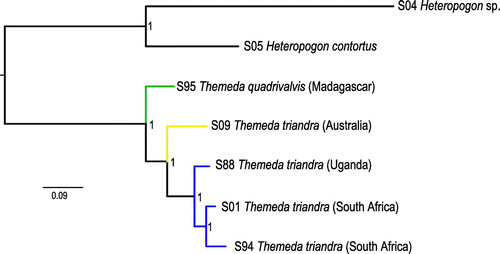
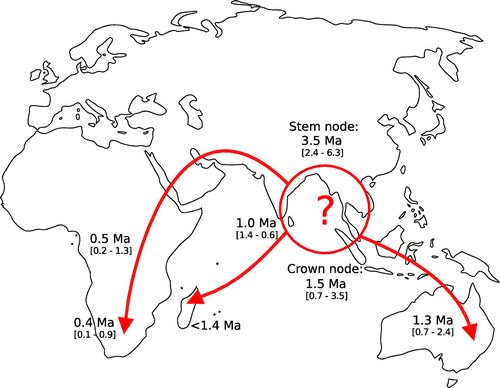
Figure 4. Inferred putative dispersal routes for Themeda. Dates are from a dated BEAST phylogeny based on chloroplast markers (Figure ).