Figures & data
Figure 2. Bayesian phylogram (95% majority rule consensus tree) of the focal Ferrissia spp. using the mitochondrial cytochrome oxidase 1 (COI) dataset. Ferrissia rivularis and Pettancylus sp. were used as outgroups. Node statistical support is reported as nodal posterior probabilities (Bayesian Inference of phylogeny, BI) and bootstrap values (Maximum Likelihood, ML). Asterisks indicate a bootstrap support value lower than 50. •: Novel sequences.
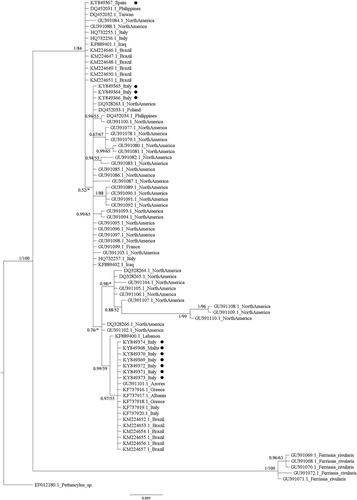
Figure 3. Bayesian phylogram (95% majority rule consensus tree) of the focal Ferrissia spp. using the mitochondrial 16S ribosomal subunit (16S rRNA) dataset. Ferrissia rivularis and Pettancylus sp. were used as outgroups. Node statistical support is reported as nodal posterior probabilities (Bayesian Inference of phylogeny, BI) and bootstrap values (Maximum Likelihood, ML). Asterisks indicate a bootstrap support value lower than 50. •: Novel sequences.
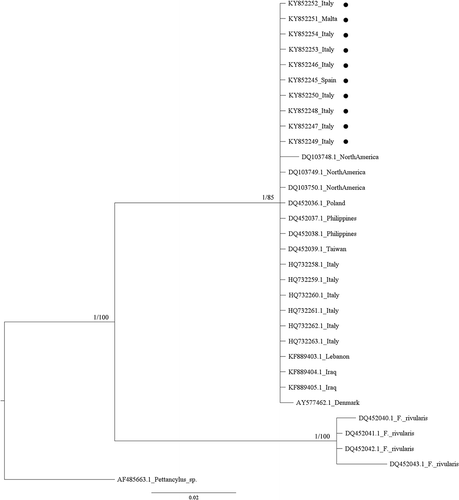
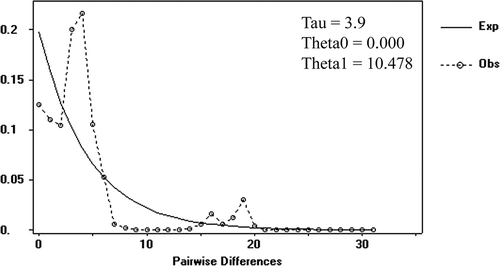
Figure 4. Median-joining haplotype network based on a fragment of mtDNA cytochrome oxidase sub-unit 1 (COI). Dashes indicate substitution steps.
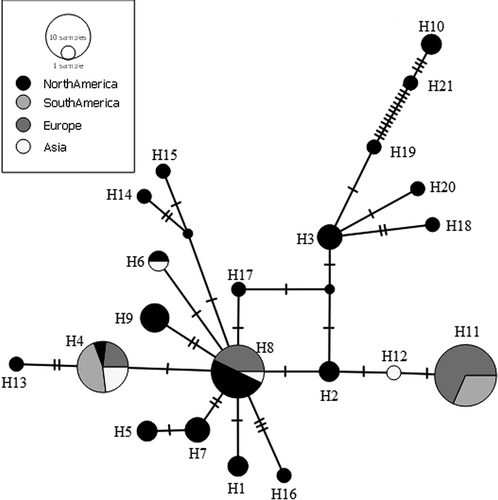
Table I. Nucleotide diversity (Pi) and haplotype diversity (Hd) of the “autochthonous” and “allochthonous” Ferrissia groups based on the mitochondrial cytochrome oxidase 1 (COI) and 16S ribosomal subunit (16S rRNA) gene fragments.
Table II. AMOVA FST results based on 578-bp-long fragments of the mtDNA COI gene. “Va” is the variance among groups, “Vb” is the variance among populations within groups.