Figures & data
Figure 1. Area of study. Red and blue dots refer to sampling sites of Chiloneus meridionalis and C. hoffmanni, respectively
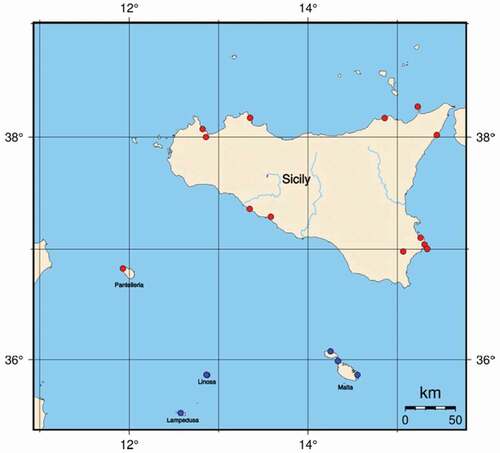
Figure 2. Habitus of (a) Chiloneus meridionalis from Palermo, Monte Pellegrino, and (b) C. hoffmanni from Malta, St. Thomas Bay
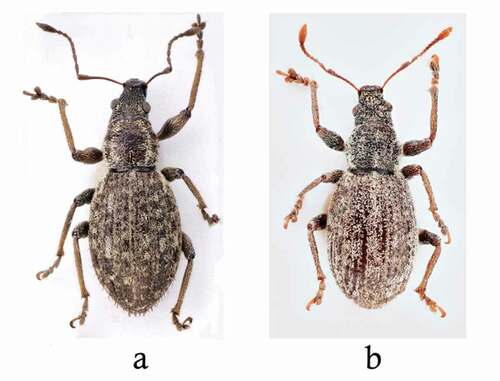
Figure 3. Left protibia of (a) C. meridionalis and (b) C. hoffmanni of the same specimens as those shown in Figure 2. Last antennomeres and club of (c) C. meridionalis and (d) C. hoffmanni. Aedeagus of C. meridionalis in (e) dorsal and (f) lateral views. Aedeagus of C. hoffmanni in (g) dorsal and (h) lateral views
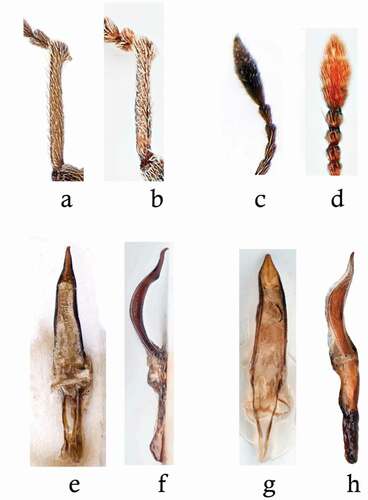
Figure 4. Bayesian consensus tree of the scored haplotypes of the cox1 gene; figures at nodes are posterior probability values; vertical black bars refer to scored groups discussed in the text; vertical double arrow shows the K80 distance between groups #1 and #2
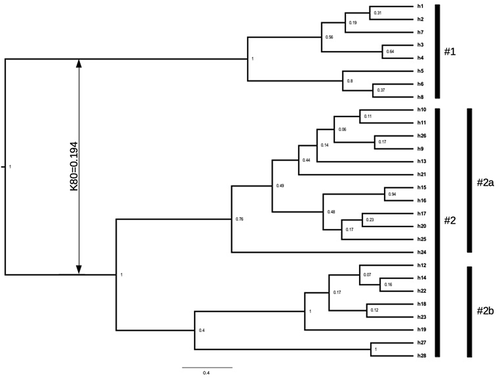
Figure 5. Statistical parsimony network of the scored cox1 haplotypes; each haplotype is represented by a circle whose size is proportional to the haplotype frequency (number of individuals found); empty circles correspond to missing or not sampled (presumed) haplotypes; numbers and abbreviations refer to groups and clades discussed in the text and illustrated in
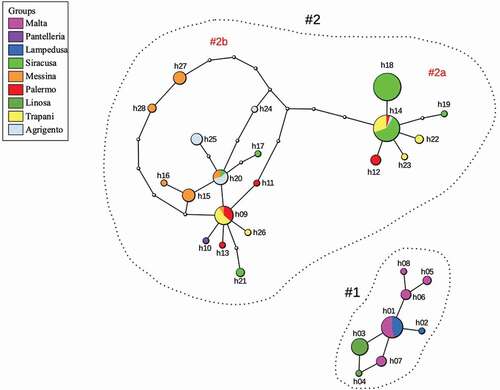
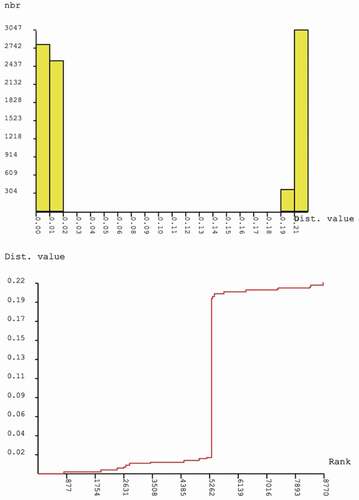
Figure 6. Results of the automatic barcoding gap discovery (ABGD) analysis using the K80 distance measure for cox1: (a) distribution of pairwise K80 distances among all samples shows two modes (low and high distance values) related to intraspecific and interspecific distances, respectively; (b) the same values plotted in ranked order show a steep slope at the barcoding gap value