Figures & data
Figure 1. Map of localities of the populations of Monacha atacis, M. samsunensis and M. cartusiana analysed (see for details).
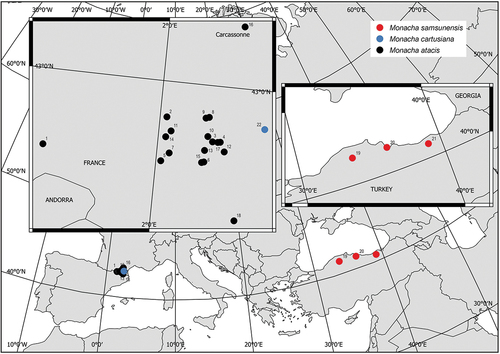
Table I. List of localities of the French and Turkish populations of Monacha atacis, M. samsunensis and M. cartusiana used for molecular and morphological research.
Table II. GenBank sequences used for molecular analysis comparisons.
Table III. Primers used in molecular analysis.
Figure 2–7. Shells of Monacha atacis from France: Grotte de Majestier [Maj3] (DCBC & MNHW-F.18.41; FGC 51099) (2), Saint-Ferriol [Fer2-1] (DCBC & MNHW-F.18.39; FGC 51097) (3) and Carcassonne (FGC 35773) (4) and Monacha samsunensis from Turkey: Kastamonu [Kas2: 5; Kas6: 6; Kas3: 7] (DCBC; FGC 51094) (5–7).
![Figure 2–7. Shells of Monacha atacis from France: Grotte de Majestier [Maj3] (DCBC & MNHW-F.18.41; FGC 51099) (2), Saint-Ferriol [Fer2-1] (DCBC & MNHW-F.18.39; FGC 51097) (3) and Carcassonne (FGC 35773) (4) and Monacha samsunensis from Turkey: Kastamonu [Kas2: 5; Kas6: 6; Kas3: 7] (DCBC; FGC 51094) (5–7).](/cms/asset/29a58005-1044-4066-9ddc-62e126757aeb/tizo_a_2100932_f0002_oc.jpg)
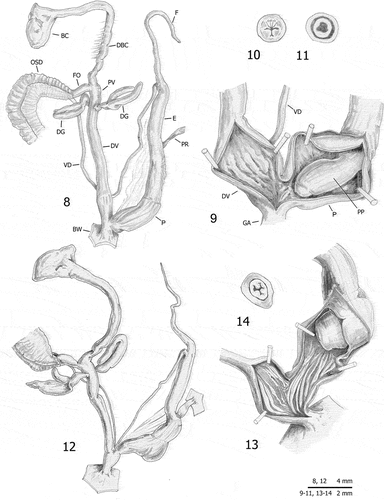
Figures 8–14. Distal genitalia (8, 12), internal structure of distal genitalia (9, 13), transverse sections of medial epiphallus (10) and apical penial papilla (11, 14) of Monacha atacis from France: Carcassonne (FGC 35773) (8–11) and Lapradelle (DCBC & MNHW-F.18.27; FGC 51247) (12–14).
Figures 15–19. Distal genitalia (15–16), internal structure of distal genitalia (17), transverse sections of medial epiphallus (18) and apical penial papilla (19) of Monacha samsunensis from Turkey: Atakum/Samsun [Bey3: 15; Bey2: 16–19] (DCBC; FGC 51175).
![Figures 15–19. Distal genitalia (15–16), internal structure of distal genitalia (17), transverse sections of medial epiphallus (18) and apical penial papilla (19) of Monacha samsunensis from Turkey: Atakum/Samsun [Bey3: 15; Bey2: 16–19] (DCBC; FGC 51175).](/cms/asset/965c0b8c-3b6b-49c1-b73c-237f7460129b/tizo_a_2100932_f0004_b.gif)
Figures 20–22. Distal genitalia (20–21) and internal structure of distal genitalia (22) of Monacha samsunensis from Turkey: Kastamonu [Kas3: 20; Sam3: 21–22] (DCBC; FGC 51094, 51095).
![Figures 20–22. Distal genitalia (20–21) and internal structure of distal genitalia (22) of Monacha samsunensis from Turkey: Kastamonu [Kas3: 20; Sam3: 21–22] (DCBC; FGC 51094, 51095).](/cms/asset/71667ab1-f305-4db5-ba85-3f83ae475ad7/tizo_a_2100932_f0005_b.gif)
Figure 23. Maximum Likelihood (ML) tree of concatenated COI + (short) 16S rDNA haplotypes obtained from specimens of Monacha atacis and Monacha samsunensis compared with sequences obtained from GenBank for representatives of the other Monacha species. Concatenated COI + 16S rDNA sequences (Table S1) were cut to 932 positions (600 bp COI + 332 bp 16S) in length. Numbers next to the branches indicate bootstrap support above 50% calculated for 1000 replicates from ML (left) and NJ (right) analysis (Felsenstein Citation1985). The tree was rooted with Trochulus hispidus concatenated sequences KY818415 + KY818541 deposited in GenBank by Neiber et al. (Citation2017).
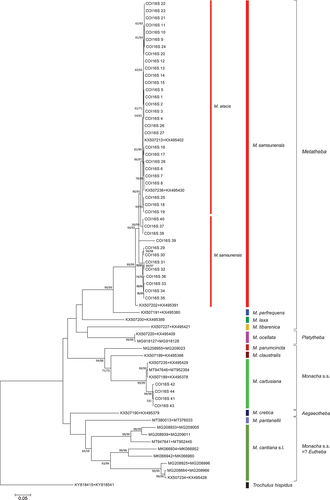
Table IV. Ranges of K2P genetic distances between analysed COI sequences (mean value in parenthesis).
Figure 24. Maximum Likelihood (ML) tree of ITS2 common sequences obtained from specimens of Monacha atacis and Monacha samsunensis compared with sequences obtained from GenBank for representatives of the other Monacha species. ITS2 sequences were cut to 564 positions in length. Numbers next to the branches indicate bootstrap support above 50% calculated for 1000 replicates from ML (left) and NJ (right) analysis (Felsenstein Citation1985). The tree was rooted with Trochulus hispidus ITS2 sequences KX495451 and MG585474 deposited in GenBank by Neiber and Hausdorf (Citation2017) and Caro et al. (Citation2019), respectively.
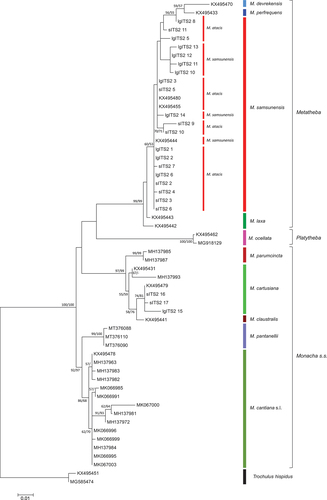
Figure 25. Maximum Likelihood (ML) tree of concatenated ITS2 + H3 common sequences obtained from specimens of Monacha atacis and Monacha samsunensis compared with sequences obtained from GenBank for representatives of the other Monacha species. Concatenated ITS2 + H3 sequences (Table S2) were cut to 825 positions (546 positions ITS2 and 279 positions H3) in length. Numbers next to the branches indicate bootstrap support above 50% calculated for 1000 replicates from ML (left) and NJ (right) analysis (Felsenstein Citation1985). The tree was rooted with Trochulus hispidus ITS2 + H3 concatenated sequences MT755395 and MT758614 deposited in GenBank by Proćków et al. (Citation2021).
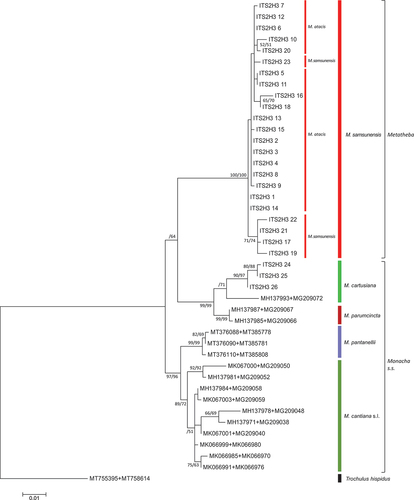
Figure 26. Maximum Likelihood (ML) tree of concatenated COI + 16S rDNA + (5.8S rDNA + ITS2 + 28S rDNA) sequences of Monacha atacis and Monacha samsunensis compared with sequences obtained from GenBank for representatives of the other Monacha species. Concatenated sequences (Table S3) were 2325 positions in length (600 COI + 869 16S rDNA + 42 5.8S rDNA + 557 ITS2 + 257 28S rDNA). Bootstrap support above 50% from ML (left) and NJ (middle) analysis as well as posterior probabilities (right) from Bayesian inference analysis are marked at the nodes. Bootstrap analysis was run with 1000 replicates (Felsenstein Citation1985). The tree was rooted with Trochulus hispidus COI + 16S rDNA + (5.8S rDNA + ITS2 + 28S rDNA) concatenated sequences KY818415 + KY818541 + KY818647 deposited in GenBank by Neiber et al. (Citation2017).
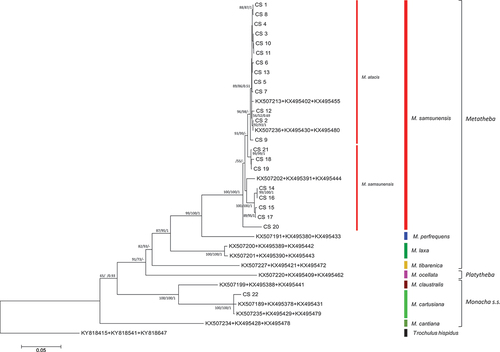
Figure 27. Shell of the Monacha samsunensis lectotype designated by Hausdorf (Citation2000a), kept in the Naturhistorischen Museum Wien (© NHMW, the inventory number: NHMW-MO-79000/K/19263) (photo by Sara Schnedl obtained by courtesy of Anita Eschner, NHMW). Scale bar 5 mm.

Table V. Ranges of K2P genetic distances between COI sequences of M. atacis and M. samsunensis populations (mean value in parenthesis).
