Figures & data
Table I. Chromosome morphometric parameters of the studied samples of H. hippocrepis and M. monspessulanus. RL = Relative length (Chromosome length/total chromosome length*100); CI = Centromeric index (short arm length/chromosome length*100); m = metacentric; sm = submetacentric; t = telocentric.
Figure 1. Karyograms (a, b, f, g and h) and metaphase plates (c-e) of H. hippocrepis stained with Giemsa (a and b), Ag-NOR (c) (male), CMA3 /MG (d) (female), NOR-FISH (e) (female) and sequential C-banding + Giemsa (f) + CMA3 (g) + DAPI (h). Arrows indicate NOR-bearing chromosomes.
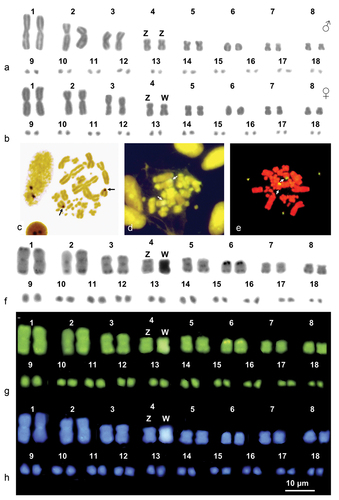
Figure 2. Karyograms (a, b, f, g and h) and metaphase plates (c-e) of M. monspessulanus stained with Giemsa (a, and b), Ag-NOR (c) (male), CMA3 /MG (d) (male), NOR-FISH (E) (female) and sequential C-banding + Giemsa (f) + CMA3 (g) + DAPI (h). Arrows indicate NOR-bearing chromosomes.
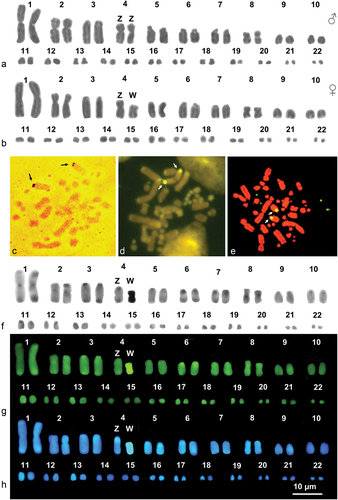
Figure 3. Phylogenetic relationships of Mediterranean whip snakes with available relative karyological data modified from .Pyron et al. (Citation2013), Figueroa et al. (Citation2016) and Zaher et al. (Citation2019). Chromosome data were collected from: (a) Mezzasalma et al. (Citation2015, Citation2018); (b) Matthey (Citation1931); (c) Gorman (Citation1973); (d) Branch (Citation1980); (e) this study; (f) Abd Allah (Citation2010)
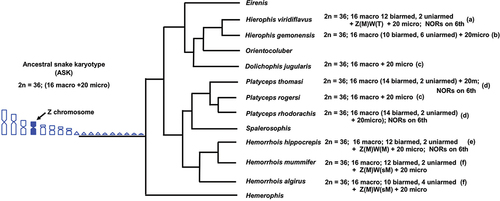
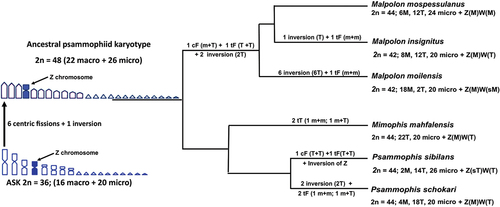
Figure 4. Phylogenetic relationships among Malpolon, Mimophis and Psammophis redrew from Pyron et al. (Citation2013), Figueroa et al. (Citation2016) and Zaher et al. (Citation2019), applied to available karyological data and the most parsimonious chromosome rearrangements occurred during specific diversification. cF = centric fusion; tF = tandem fusion; M = macrochromosome; m = microchromosome; T = telocentric macrochromosomes; sT = subtelocentric.