Figures & data
Figure 1. Two small Leuciscidae species occurring in the vicinity of Rafina in Attika. Adult Pelasgus marathonicus from the Boeotian population, Kifissos River basin (photo courtesy of J. Freyhof) (top), and Ladigesocypris ghigii from the Rafina translocated population; a very large individual: 95 mm in total length (photo by D. Zogaris) (bottom).

Table I. List of Ladigesocypris specimens, and species used for the genetic analyses. For published sequences, the original publication is indicated. (A) Durand et al. (Citation2002), (B) Freyhof et al. (Citation2006), (C) Perea et al. (Citation2010), (D) Durand et al. (Citation2000), (E) Schönhuth et al. (Citation2018), (F) Viñuela Rodríguez et al. (Citation2021), (G) Benovics et al. (Citation2023), (H) Benovics et al. (Citation2020), (J) Nejat et al. (Citation2023), * - all 32 Pelasgus spp. RAG1 haplotypes (MZ230959–MZ230990) published in Viñuela Rodríguez et al. (Citation2021) were used.
Figure 2. The 50% majority rule consensus Bayesian phylogenetic reconstructions for both mitochondrial cytochrome b and nuclear RAG1 sequence data. Numbers on branches represent posterior probabilities; only values ≥ 0.7 are shown. Samples from the introduced population near Rafina are in red.
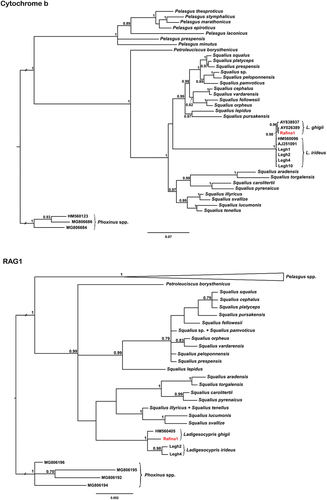
Figure 3. Haplotype networks of Ladigesocypris sequences constructed by a statistical parsimony method for both mitochondrial cytochrome b and nuclear RAG1. For RAG1, all four possible alleles of heterozygotic sample Legh4 are depicted. Arrows in the RAG1 network indicate haplotypes of Legh4 sample with the highest probability as estimated by phasing the dataset by DnaSP (probability 0.65). Intermediate non-occurring haplotypes between our haplotypes are shown as small white circles. A line between two neighbouring haplotypes indicates one mutational step.
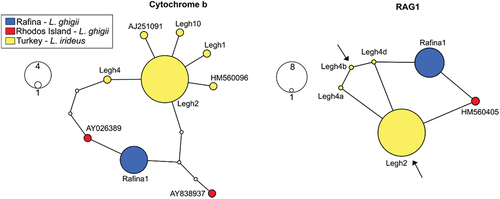
Table II. Morphometric data of Ladigesocypris ghigii found at two sites near Rafina (N = 10). SD is standard deviation.
Table III. Frequency of occurrence of meristic characters of Ladigesocypris ghigii found at two sites near Rafina (N = 10). Counts of transverse scales and transverse scales at caudal peduncle do not include the scale row on dorsal and ventral midlines noted as “1/2” in the text.
Table IV. Scoring output from the Aquatic Species Invasiveness Screening Kit for Ladigesocypris ghigii in the risk assessment area of Attika region, Greece.
