Figures & data
Figure 1. Induction of T cell response by HLA-A24 restricted SARS-CoV-2 Spike protein. (A) List of candidate peptides carrying the HLA-A24 binding motif. Fifteen peptides carrying the HLA-A24 binding motif were designed, with each peptide pool containing 5 peptides. (B) PBMC stimulation schema. PBMCs were stimulated using each peptide pool on day 0 and day 7, and the specificity of PBMCs was evaluated using each peptide-pulsed target cells. (C) IFNγ ELISpot assay. PBMCs were evaluated for peptide specificity by an IFNγ ELISpot assay. Fifty thousand PBMCs were co-cultured with each peptide-pulsed target cells, and IFNγ production was assessed. Peptide-unpulsed targets (-) were used as a negative control.
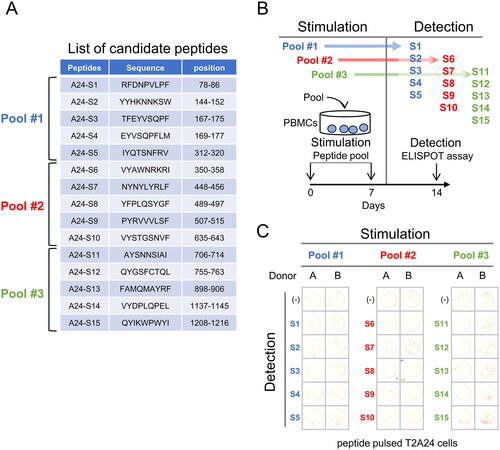
Figure 2. Establishment of NF9-specific CTL clone. (A) IFNγ ELISpot assay. PBMCs were evaluated for peptide specificity by an IFNγ ELISpot assay using NF9-peptide-pulsed T2-A24 cells as targets. Peptide-unpulsed T2-A24 cells served as a negative control. (B) FACS analysis using NF9 tetramer. PBMCs were stained with the HLA-A24 NF9 complex tetramer and analyzed by flow cytometry. The HLA-A24 HIV peptide complex tetramer was used as a negative control. NF9 tetramer-positive cells were sorted individually. (C) IFNγ ELISpot assay. Tetramer-positive clone cells were evaluated by an IFNγ ELISpot assay using NF9-peptide-pulsed T2-A24 cells as targets. Peptide-unpulsed T2-A24 cells served as a negative control.
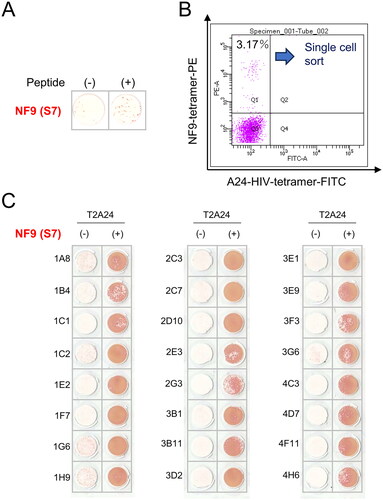
Figure 3. NF9 peptide-specific CTL clone cross-reacts with the Y453F peptide. (A) IFNγ ELISpot assay. The NF9 peptide-specific CTL clone was evaluated using NF9 peptide, L452R peptide, and Y453F peptide. Each peptide was pulsed onto T2-A24 cells, and the reactivity of the CTL clone was examined by an IFNγ ELISpot assay. Peptide-unpulsed T2-A24 cells were used as a negative control. (B) Peptide titration. Serially diluted NF9 peptide and Y453F peptide were pulsed onto T2-A24 cells and used for an IFNγ ELISpot assay. The NF9 peptide-specific CTL clone was used as effector cells.
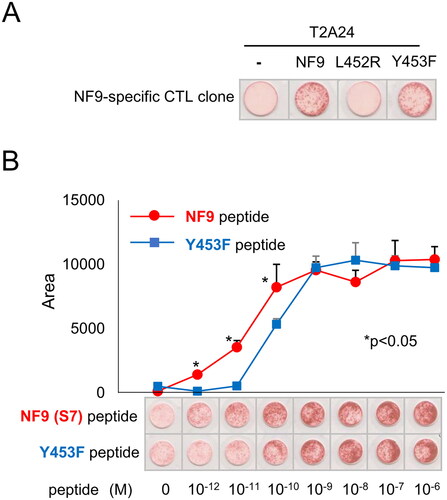
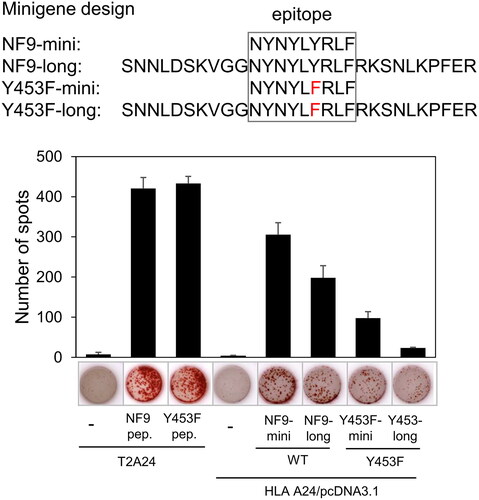
Figure 4. NF9 peptide-specific CTL clone recognizes endogenously expressed Y453F peptide. The design of minigenes (upper panel). NF9-mini encodes the minimal epitope, NF9-long encodes the epitope with 5’- and 3’-flanking sequences, Y453F-mini encodes the minimal epitope of the Y453F mutant, and Y453F-long encodes the Y453F epitope with 5’- and 3’-flanking sequences. IFNγ ELISpot assay (lower panel). Minigenes were co-transfected with HLA-a*24:02 cDNA into 293 T cells and used for an IFNγ ELISpot assay. The NF9 peptide-specific CTL clone was used as an effector. Minigene-negative 293 T cells were used as a negative control. NF9 peptide and Y453F peptide-pulsed T2-A24 cells were used as positive controls.