Figures & data
Figure 1. Inhibition of mycelial growth of Fusarium sp. strain SQUCC-F1-2 by Neocosmospora sp. strain SQUCC-F1-1 and Alternaria alternata strain SQUCC-F6-2. A, Alternaria alternata; N, Neocosmospora sp.; F, Fusarium sp.
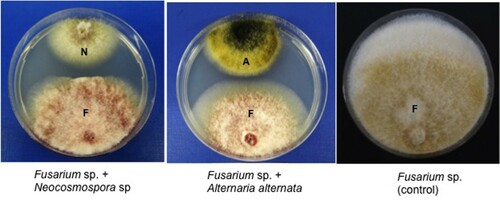
Figure 2. Scanning electron micrographs showing morphological changes in the hyphae of Fusarium sp. strain SQUCC-F1-2 due to antagonistic activity of Alternaria alternata strain SQUCC-F6-2 and Neocosmospora sp. strain SQUCC-F1-1. A, Fusarium sp. control; B, Fusarium sp. + Alternaria alternata; C, Fusarium sp.+ Neocosmospora sp.
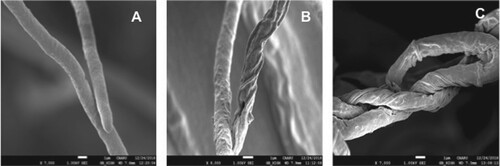
Figure 3. Phylogenetic tree obtained from the combined ITS and Alt a 1 sequence alignment of the species of Alternaria. The tree was rooted using Alternaria alternantherae (CBS 124392). The phylogenetic tree was constructed by Maximum likelihood methods and GTRGAMMA model. The bootstrap values are expressed as percentage of 1000 replications.
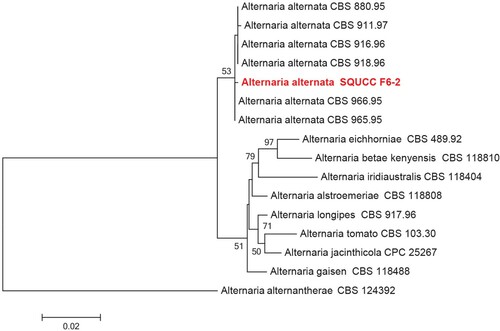
Figure 4. Phylogenetic tree obtained from combined ITS, TEF and RPB2 sequence alignment of the species of Neocosmospora. The tree was rooted using Geejayesia atrofusca (NRRL 22316) and Geejayesia cicatricum (CBS 125552). The phylogenetic tree was constructed by Maximum likelihood methods and GTRGAMMA model. The bootstrap values are expressed as percentage of 1000 replications.
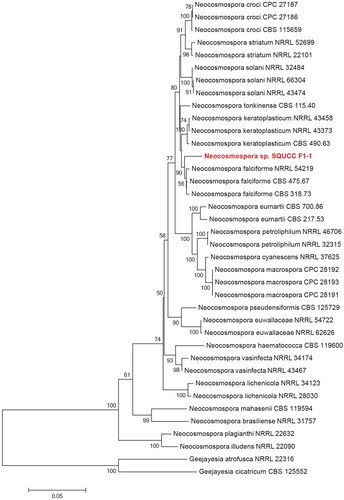
Table 1. Compounds detected in the extract of Neocosmospora sp. strain SQUCC-F1-1.
Table 2. Compounds detected in the extract of Alternaria alternata strain SQUCC-F6-2.
Data Availability
All data generated or analyzed during this study are included in this article.
