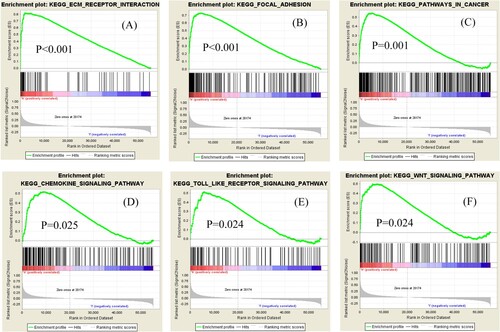
Figures & data
Figure 1. LUM differential expression in normal and tumor tissues. A: comparison of TCGA profile; B: comparison of GEO dataset
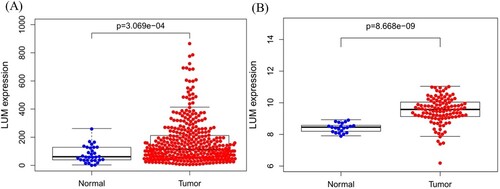
Figure 2. Validation of protein expression of LUM in normal tissue (A) and gastric cancer (B) using the Human Protein Atlas database.
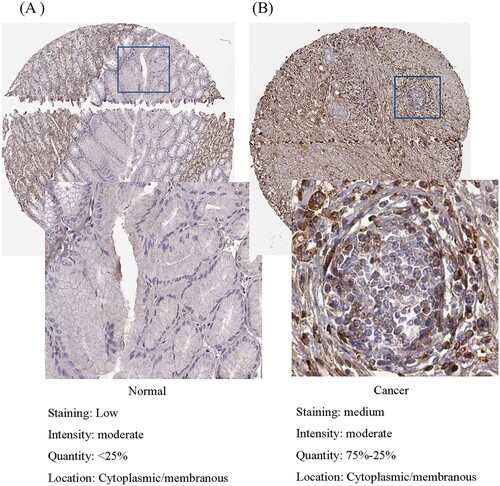
Figure 3. Association of LUM expression with clinical variates. A: age; B: gender; C: grade; D: distant metastasis; E: lymph node metastasis; F: clinical stage; G: depth of invasion
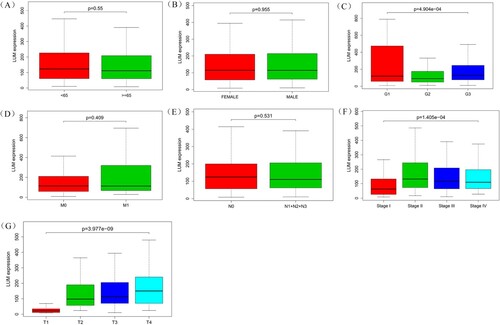
Table 1. Logistic regression between LUM expression and clinicopathologic parameters.
Figure 4. Receiver operating characteristic curve for LUM expression in normal and gastric cancer tissues.
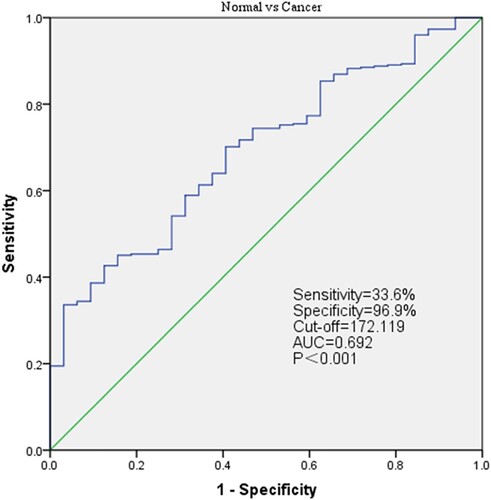
Figure 5. LUM expression and overall survival in gastric cancer patients in TCGA profile (A) and the GSE84437 dataset (B).
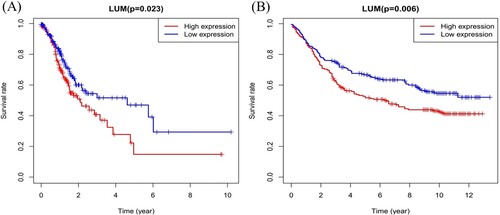
Table 2. Univariate and multivariate analysis of the correlation between LUM expression and overall survival in gastric cancer patients.
Table 3. Univariate and multivariate analysis of the correlation between LUM expression and overall survival in gastric cancer patients validated using the GSE8443 dataset.