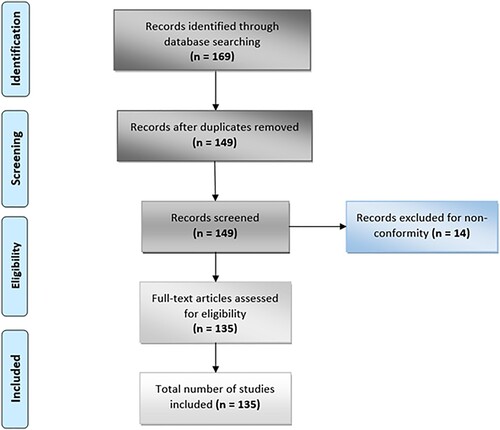
Figures & data
Figure 2. An illustration of the genomic organization and structure of SARS-CoV-2. (A) Genomic organization of SARS-CoV-2. The ORF1a and ORF1b of the viral RNA genome encode 16 non-structural proteins (nsp1–nsp16). Nsp3 and nsp5 encode PLpro and 3CLpro, respectively, which are essential for viral replication and pathogenesis. The genomic organisation of S glycoprotein showing its two subunits (S1 and S1) is depicted below the viral genome. Dotted lines within this structure indicate the S1/S2 cleavage site. Other important components of the S glycoprotein such as signal protein (SP), receptor-binding domain (RBD), fusion peptide (FP), heptad repeat (HR1 and HR2), transmembrane domain (TM), and cytoplasm domain (CP) are depicted. (B) Structure of SARS-CoV-2 showing its single-stranded positive-sense RNA genome and essential structural proteins.
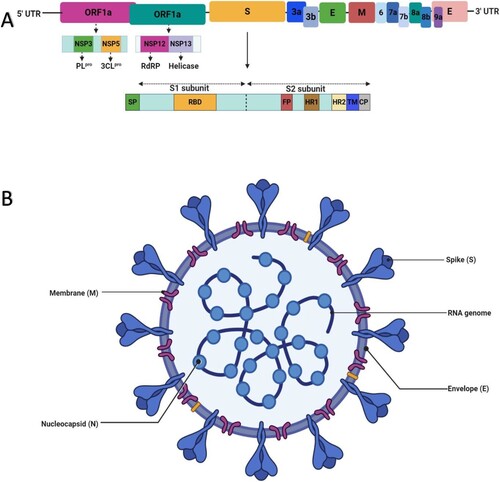
Table 1. List of 16 coronaviral non-structural proteins with their functions.
Table 2. Compounds with potential against SARS-CoV-2 non-structural proteins evaluated through computational screening.
Table 3. Virtually screened phytocompounds with activity against SARS-CoV-2 structural proteins and host-associated proteins.
Figure 4. Interaction of representatives of phytoligands, quercetin, and tryptanthrin with SARS-CoV-2 3CLpro and PLpro, respectively (Mani et al. Citation2020) (Reuse permission was obtained from Elsevier, Licence number: 5034780990710).
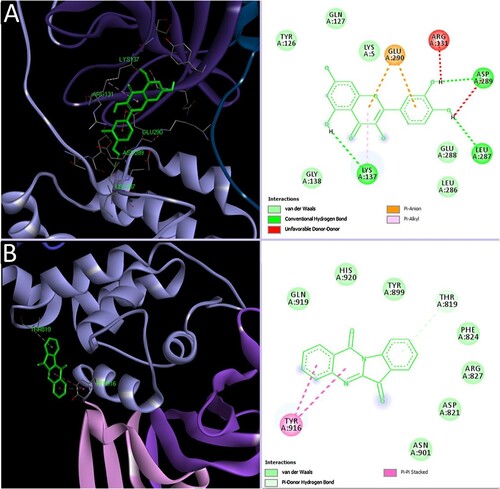
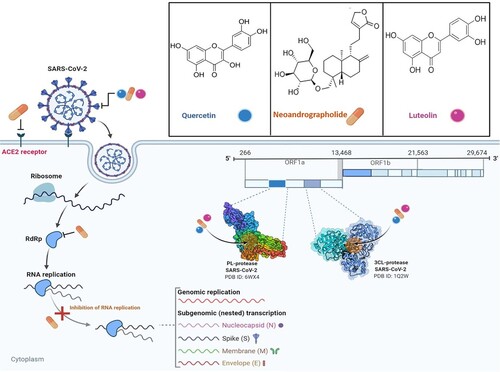
Figure 5. The life cycle of SARS-CoV-2 and potential druggable targets of representative phytocompounds. Quercetin and luteolin predictably interact with 3CLpro, PLpro, and S glycoprotein, while neoandrographolide acts against 3CLpro, PLpro, host ACE2 receptor, RdRp and may also inhibit RNA replication.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.