Figures & data
Table 1. General data of pyrophosphate sequencing.
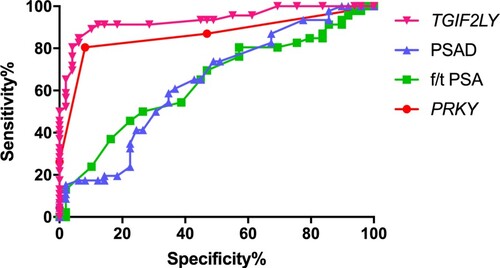
Figure 1. Bioinformatics analysis of PRKY and TGIF2LY expression in prostate cancer. The expression of PRKY was high in cancer tissues and low in normal tissues, and the difference was statistically significant (P < 0.001, Figure (A)). The expression of TGIF2LY in prostate cancer tissues was significantly lower than that in normal tissues (P < 0.001, Figure (B)).
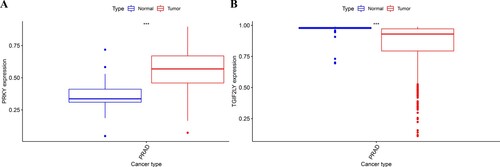
Table 2. The clinical characteristics of the two groups of patients.
Data availability statement
All the original data of this study can be accessed from https://figshare.com/s/b2786bd44a844ba9b9c5. Publicly available data was also obtained from the UCSC Xena (https://xena.ucsc.edu/) TCGA database, reference [GDC TCGA Prostate Cancer (PRAD)].