Figures & data
Figure 1. Pedigree structure of the Chinese family. Family members with disease are indicated by the shading. Squares and circles denote males and females, respectively. Roman numerals indicate generations. The arrow indicates the proband (III-1).
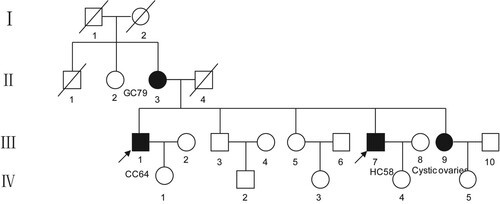
Table 1. The clinical characteristics of four patients in this family.
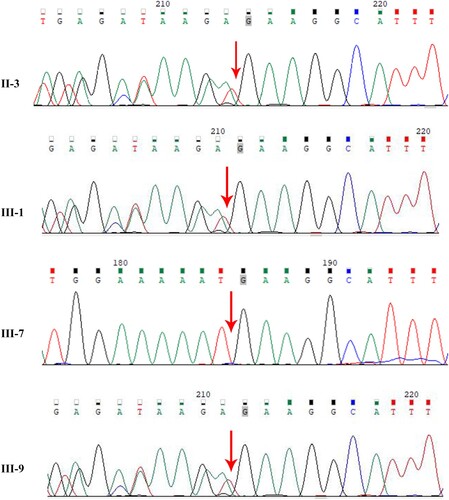
Figure 2. Identification of this truncation mutation c.7141_7151del in ATM. Visualization of ATM sequencing reads containing c.7141_7151del mutation using the Integrative Genomics Viewer (IGV). The big red box represents the 11 missing nucleotides. The small red box represents the stop codon.
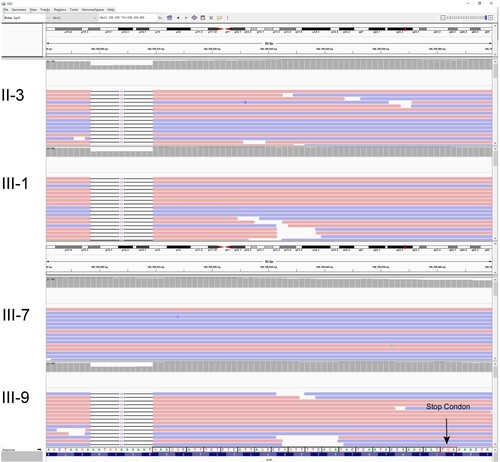
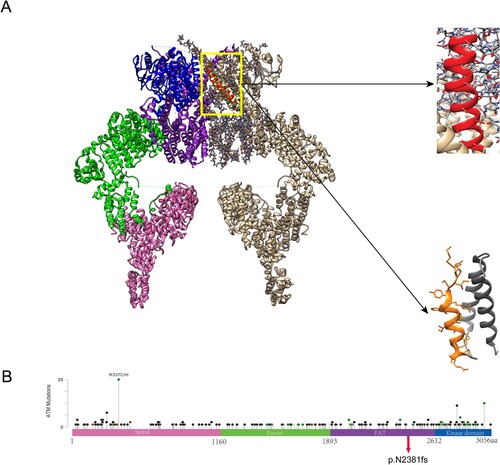
Figure 3. Mutations in ATM. (A) Protein structure of ATM dimer. On the left is the normal ATM protein structure, which serves as a control. The pink part represents the spiral region, the green part is the pincer region, the purple is the FAT structure region, and the blue is the kinase domain. On the right, the gray part is the unexpressed part after ATM mutation. The enlarged red part represents the unshifted region of a normal ATM protein. The enlarged yellow portion represents the amino acid sequence encoded by ATM after a code-shift due to deletion mutations. (B) The circle represents the mutation site, and the height represents the mutation frequency. ATM proteins are showing the spiral region, pincer region, FAT region, and kinase domain. Mutation p.N2381fs was found in the kinase domain of ATM.
Data availability statement
The data supporting the findings of this study are available in the figshare repository [https://figshare.com/] at doi:10.6084/m9.figshare.19709965 [https://figshare.com/s/3e18591f29c0c9c8c397].