Figures & data
Table 1. The sequences of primers used in this study.
Table 2. Main characteristics of the participants.
Table 3. Status of Helicobacter pylori infection among participants of different main characteristics.
Table 4. Association analysis between IL4, TNFA and IL10 genes polymorphisms and H. pylori infection.
Figure 1. LD analysis of the candidate gene SNPs. The results of LD analysis for IL4 (A), TNFA (B) and IL10 (C) genes were described. The color shift from white to red represented that the results of D’ and r2 changed from 0 to 1. A deep red block means the result is 1. The closer the value is to 1, the higher degree of LD is. Kb represented the distance between two SNPs in the genome. LD was observed from the three SNPs of TNFA gene and the two SNPs of IL10 gene, respectively.
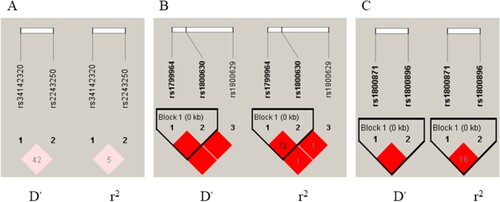
Table 5. Results of haplotype analysis.
Data availability statement
The data that support the findings of this study are openly available in figshare at https://doi.org/10.6084/m9.figshare.21563523.
