Figures & data
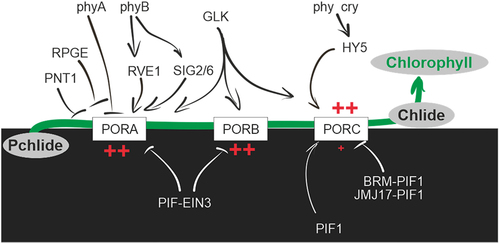
Figure 1. Photoreceptors in higher plants. The graph illustrates the absorption spectra of a plant extract due to the presence of chlorophyll a, b, and carotenoids. The photoreceptors responsible for perceiving different light qualities and their subsequent physiological effects on photosynthesis are also highlighted.
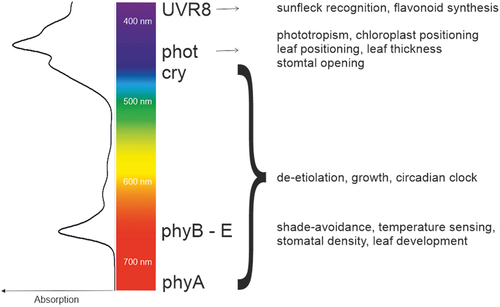
Figure 2. Schematic view of the regulation of the expression of the plastid encoded genes by photoreceptors in the dark and the light. The transition from the heterotrophic (dark) to the photoautotrophic (in the light) state is depicted left to right. Nuclear-encoded NEP induces the expression of housekeeping genes and PEP. In the light, nuclear-encoded factors assist in assembling the PEP complex leading to the formation of functional photosynthetic complexes. The colour code specifies the photoreceptors involved in regulating the nuclear-encoded factors. N, nucleus; CP, chloroplast; PS, Photosynthesis. Please refer to the text for further details.
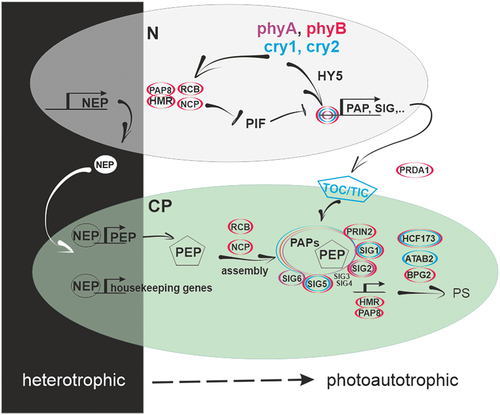
Figure 3. Schematic model of the dynamics of the core factors needed to regulate PhANG expression in the nucleus during darkness, transitional periods and in the light. The triangles depict varying levels of active protein found within the cell. Proteins coded in blue stabilise while those coded in red lead to activation or inactivation or degradation. Expression regulators are shown in a square, and interacting regulators are shown in a circle. The asterisk (*) denotes activated photoreceptors. Please refer to the text for further details.
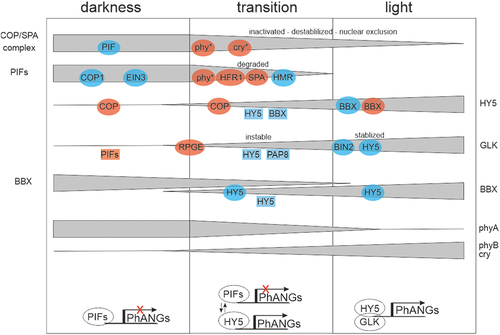
Figure 4. Schematic view of the regulation of the three POR genes present in Arabidopsis thaliana. PORA, B, and C catalyse the synthesis of Chlide from Pchlide, which is the light-dependent step in chlorophyll biosynthesis. Factors that activate or repress in the dark (bottom) and light (top) are indicated. Arrows and bars represent positive and negative regulation, respectively. The position of the + represents the light condition in which the respective POR genes are highly expressed. Please refer to the text for further details.