Figures & data
Table 1. PCR Primers Used for RT‐PCR
Figure 1. Purity of the extracted RNA. RNA from lung tissue frozen in Optimal Cutting Temperature compound was analyzed by UV absorbance between 220 and 300 nm. (A) RNA extracted with RNeasy Mini Kit. (B) RNA extracted with TRIzol reagent. Both show single peaks at 260 nm.
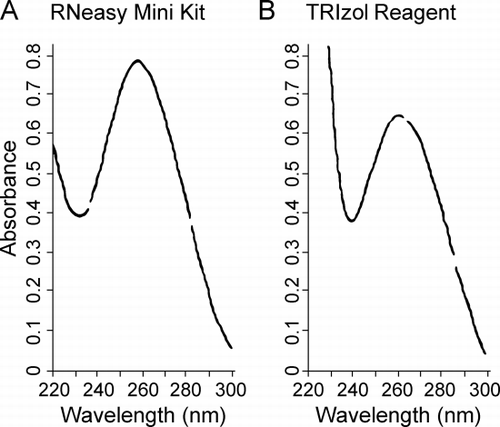
Figure 2. RNA size analysis. RNA was analyzed on the Agilent 2100 Bioanalyzer. (A) Representative results of the analysis displayed in a gel electrophoresis format. RNA extracted with TRIzol from human bronchial epithelial (HBE) cells (lane C), frozen tissues (lanes F1–F4), and paraffin‐embedded tissues (lanes P1–P2). Lane M is an RNA ladder as size markers with the nucleotide sizes on the left and the expected positions of the 28 and 18S ribosomal RNA indicated on the right of the gel. (B) Representative results displayed in chromatographic format. Panels are labeled as the lanes in (A) and correspond to the same RNA samples as in (A), except F3 and F4 are not shown.
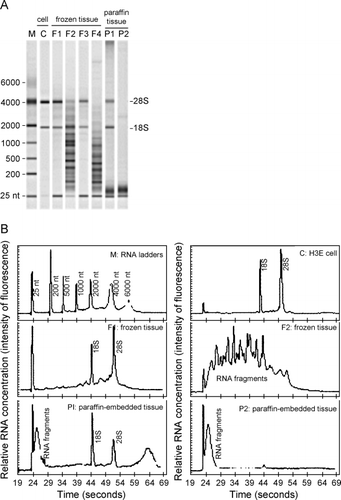
Figure 3. Confirmation of RT‐PCR products by Southern hybridization. (A) Agarose gel of the 375‐bp actin RT‐PCR product amplified from RNA extracted from A549 cells, frozen lung tissue and paraffin‐embedded lung tissue and, as positive and negative controls of hybridization, a partial restriction digest of pBS‐actin (lane +) and the purified restriction fragment carrying the sequence confirmed glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) cDNA from pBS‐GAPDH (lane −), respectively. (B) Same as in A except the 296‐bp GAPDH product was amplified and controls are the GAPDH cDNA from pBS‐GAPDH (lane +) and the purified restriction fragment carrying the sequence confirmed actin cDNA from pBS‐actin (lane −). (C) Film detecting chemiluminescence after Southern transfer of (A) and hybridization with the actin cDNA from pBS‐actin used above. (D) Same as in (C) except the gel in (B) was transferred followed by hybridization with cDNA from pBS‐GAPDH used above. Lane M is a 100‐bp DNA ladder with bp sizes indicated at the left of (A).
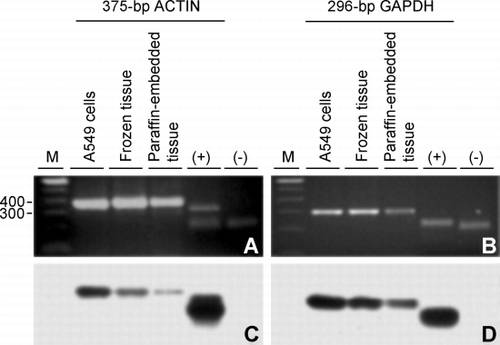
Figure 4. Integrity of RNA extracted from frozen versus paraffin‐embedded tissue. RNA extracted from (C) A549 cells, (F) frozen tissue, and (P) paraffin‐embedded tissue was analyzed by RT‐PCR using primer pairs specifying the 375‐bp actin, 296‐bp glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) and 983‐bp GAPDH targets. Ethidium bromide‐stained gels of the 375‐bp actin (lanes 2–5), 296‐bp GAPDH (lanes 7–10) and 983‐bp GAPDH (lanes 12–15, 16, 17) products are shown and include the respective negative controls (−) where the cDNA was replaced by distilled water in the PCR reactions. cDNA was synthesized from equal amounts of RNA as template using oligo (dT)25 as the primer for reverse transcriptase except for lane 17 where random hexamers were used. (M) 100‐bp DNA ladders with bp sizes indicated at the left of the gel for the 375‐bp actin and 296‐bp GAPDH targets and at the right for the 983‐bp GAPDH target.
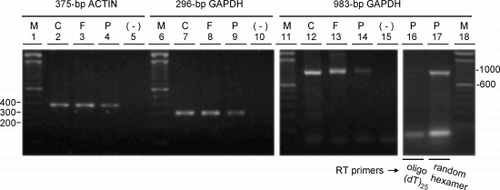
Figure 5. RNA integrity versus tissue storage time. Ethidium bromide‐stained agarose gels of (A) the 296‐bp glyceraldehyde 3‐phosphate dehydrogenase (GAPDH), (B) 983‐bp GAPDH and (C) 499‐bp integrin‐linked kinase (ILK) RT‐PCR products amplified from equal amounts of RNA extracted from frozen lung tissues that were selected to span entry dates into the tissue bank between 1995 to 2001. (−) negative control where cDNA was replaced with distilled water in the PCR reactions; (M) size markers with the corresponding bp sizes indicated at the left of the gels.
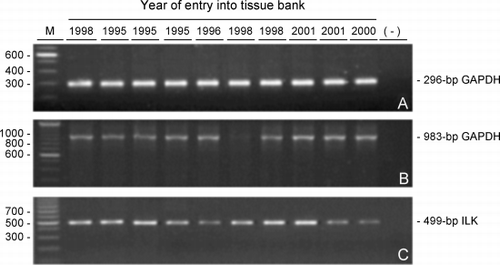
Figure 6. Minimal requirement of total RNA for RT‐PCR. Initial RNA concentrations were determined by the Agilent 2100 Bioanalyzer. Ethidium bromide‐stained agarose gels of the 296‐bp glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) product from RNA from A549 cells (C1) and from the samples shown in representing each of the two different tissue sources, frozen tissue with (F1) or without (F2) ribosomal RNA subunits and paraffin‐embedded tissue with (P1) or without (P2) ribosomal RNA subunits are shown where the RNA was serially diluted 5‐fold before RT‐PCR. The amount of RNA used for RT‐PCR is indicated above the gels. Similarly, RNA extracted from decreasing numbers of A549 cells (C2) was amplified. Leftmost lanes in each gel (M) are size markers with bp sizes indicated at the left of the gels.
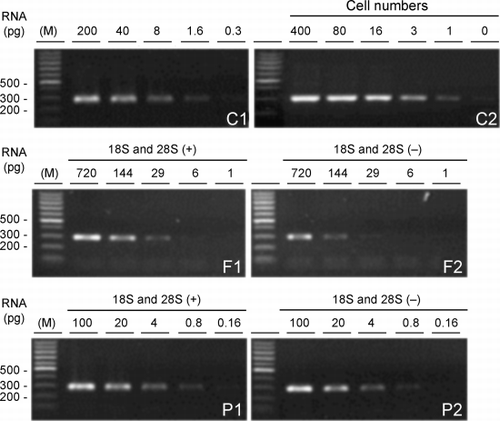
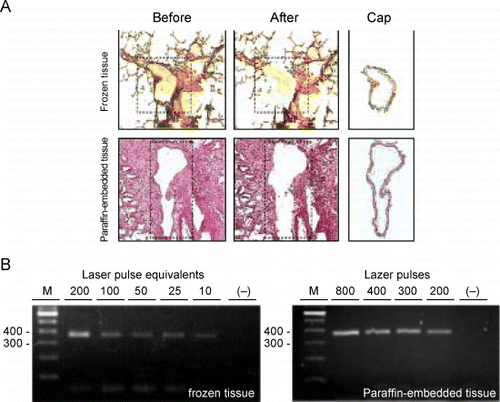
Figure 7. RT‐PCR of RNA extracted from cells obtained by laser capture microdissection (LCM). A. Representative micrographs of tissue sections stained with hematoxylin and eosin and captured cells processed by LCM. Top row represents a section of frozen lung tissue; bottom row: paraffin‐embedded lung tissue. Leftmost column represents sections before transfer of cells by LCM. The dashed line outlines the areas from which the cells will be transferred. Middle column represents the same sections after LCM. Note that the layer of airway tissue is absent. Rightmost column represents the cells that were captured onto the caps. The bar in the bottom right micrograph is representative of the magnification on all six micrographs and equals 200 µm. B. On the left is an agarose gel of the 375‐bp actin RT‐PCR products from 2‐fold serial dilutions of RNA that was extracted from airway cells captured with 200 laser pulses from frozen lung tissue and on the right, of RNA extracted from cells captured from paraffin‐embedded lung tissue using decreasing numbers of laser pulses. The lanes marked “−” represent RT‐PCR of the distilled water negative control. The leftmost lanes in each gel (M) are size markers with bp sizes indicated to their left. (Full color version available online.)