Figures & data
Table 1 List of the ammonia-oxidizing bacteria used in this study
Table 2 Nucleotide sequences of polymerase chain reaction primers
Figure 1 Partial deduced amino acid sequences of pyruvate kinase (PYK) from ammonia-oxidizing bacteria (AOB). The AOB sequences were compared to the sequences of Escherichia coli type II PYK (Ec). *Homogeneity among all the species. Functionally important amino acid residues are indicated with white letters (black background).

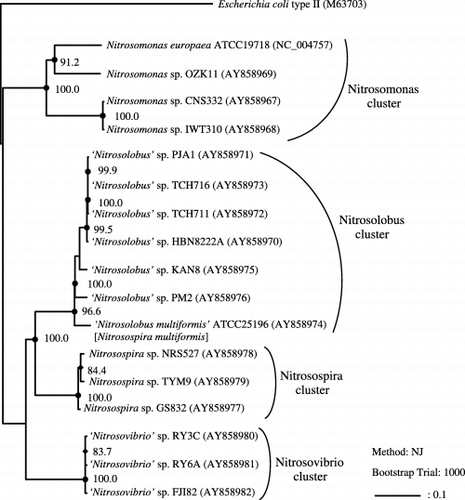
Figure 2 Phylogenetic tree inferred from comparison of pyk sequences of the β-proteobacterial ammonia-oxidizing bacteria (AOB). The tree was generated using the neighbor-joining method, and Escherichia coli type II pyk was defined as an outgroup organism. Bars represent 10% sequence divergence. Numbers indicate bootstrap values (1000 resamplings; %). Bootstrap values above 90% (•) and 70% (⧫), and accurate values are shown. Accession numbers are indicated in parentheses and based on the present officially approved nomenclature. Morphotypic and genotypic-related groups of organisms were designated as follows: Nitrosomonas cluster, Nitrosospira cluster, Nitrosolobus cluster and Nitrosovibrio cluster, and Nitrosomonas, Nitrosospira, ‘Nitrosolobus’ and ‘Nitrosovibrio’, respectively.