Figures & data
Figure 1 Location of the sampling sites of the rhizosphere soils of Miscanthus sinensis grown on acid sulfate soil (Rankoshi, Hazu, Nago) and sandy soil (Atsuma) in Japan.
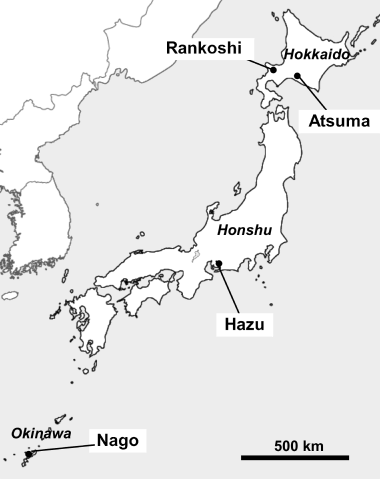
Table 1 Climates and surrounding vegetation of the experimental sites
Figure 2 Frequency distributions of (a) pH, (b) Troug-P, (c) total nitrogen and (d) total carbon of the rhizosphere soil of Miscanthus sinensis collected from Rankoshi, Hazu, Nago and Atsuma.
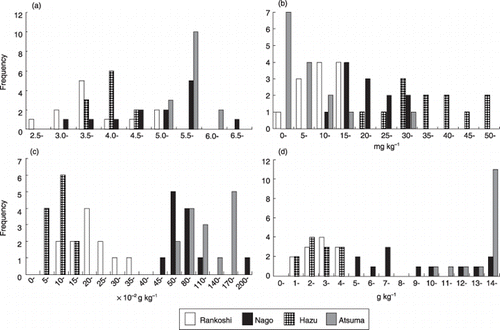
Figure 3 Phylogenetic analysis based on the sequences of a 700–760 bp fragment of arbuscular mycorrhizal (AM) fungal LSU rDNA obtained by the Miscanthus sinensis trap culture using rhizosphere soils collected from Rankoshi (purple), Hazu (blue), Nago (green) and Atsuma (orange). The neighbor-joining tree has been drawn using NJplot. Bootstrap values more than 70% are indicated. Representative sequences from each root sample are incorporated. Clone names are followed by their GenBank accession numbers for the sequences obtained in this study. The accession numbers of the reference sequences are: Glomus intraradices 107, AY639221; Glomus clarum, AJ510243; Glomus manihotis, AM158947; Glomus sp. hr11, AM040407; Glomus sp. rp2, AM040435; Glomus caledonium, AM040317; Glomus mosseae, DQ469129; Glomus etunicatum, AF145749; Scutellospora pellucida, AY639326; Scutellospora sp. hr83, AM040378; Gigaspora margarita, AF396783; Gigaspora gigantea, AY900504; Acaulospora mellea, AY900513; Acaulosporaceae sp. S175, AB206249; Acaulospora longula, AM040293; uncultured glomeromycete 6.8, AY639357; uncultured glomeromycete Cp193, AB206207; Archaeospora gerdemannii, AJ510234; Palaglomus occultum, AJ271713; Neurospora crassa, AF286411.
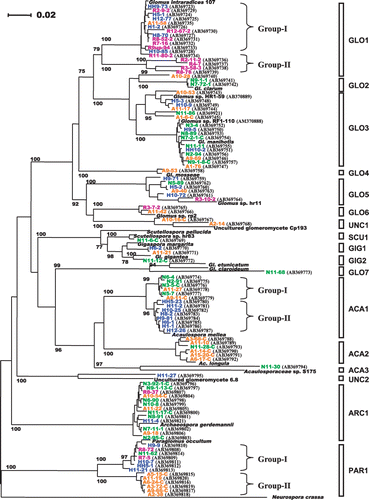
Figure 4 Scatter plot of Chao's Sørensen similarity indices of arbuscular mycorrhizal fungal communities against geographical distance between two sampling sites. R, Rankoshi; H, Hazu; N, Nago; A, Atsuma. No significant correlation between the two parameters was found (P < 0.05). Vertical bars indicate standard deviation.
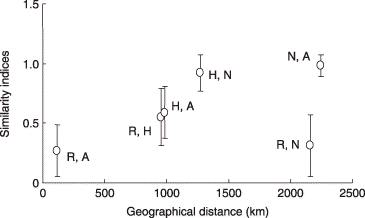
Figure 5 Principal component analysis on arbuscular mycorrhizal (AM) fungal communities at Rankoshi, Hazu, Nago and Atsum. (a) Component plot of experimental sites. The first (PC1) and second (PC2) principal components explained 40.2% and 27.1% of the total variation, respectively. (b) Scatter plot of the PC1 and PC2 scores of the AM fungal phylotypes.
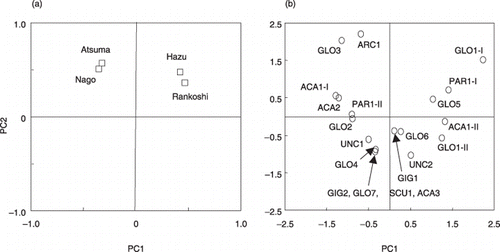
Figure 6 Differences in the pH range of soils in which arbuscular mycorrhizal (AM) fungal phylotypes occur. The pH values of rhizosphere soils from which each AM fungal phylotype was detected were plotted (○). Only phylotypes that occurred in four or more soil samples (across the all sites) were included in this analysis. Equality of variance tests showed that the datasets of ACA2 and PAR1 group-II had significantly smaller variances than those of the other phylotypes; thus, these two phylotypes were excluded from the subsequent anova. Different letters indicate significant differences in pH distribution (Tukey–Kramer test, P < 0.05). The pH values were transformed to real numbers for the calculation of mean value (▴), but were retransformed to logarithmic values for anova for normalization.
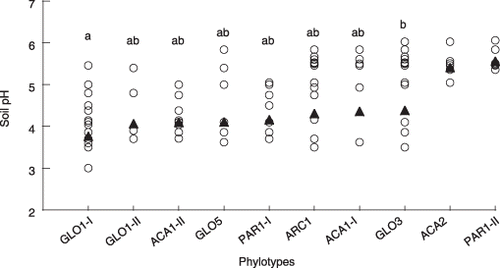
Figure 7 Scatter plot of the first principal component (PC1) scores of arbuscular mycorrhizal (AM) fungal phylotypes against the average pH values of the soils in which they occurred. Only phylotypes detected from four or more soil samples (across the all sites) were included in this analysis. The phylotypes are identified by numbers as follows: 1, GLO1 group-I; 2, GLO1 group-II; 3, ACA1 group-II; 4, GLO5; 5, PAR1 group-I; 6, ARC1; 7, ACA1 group-I; 8, GLO3; 9, ACA2; 10, PAR1 group-II.
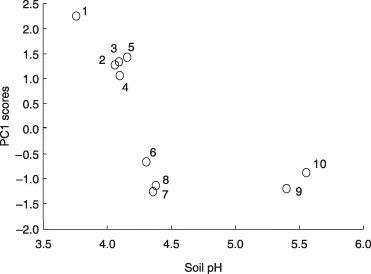
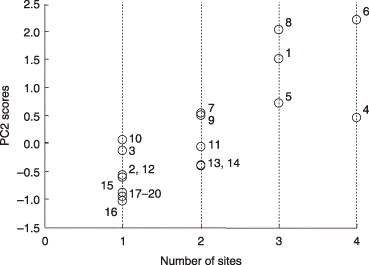
Figure 8 Scatter plot of the second principal component (PC2) scores of the phylotypes against the number of sites at which they were detected. All phylotypes were included in this analysis. The phylotypes are identified by numbers as follows: 1, GLO1 group-I; 2, GLO1 group-II; 3, ACA1 group-II; 4, GLO5; 5, PAR1 group-I; 6, ARC1; 7, ACA1 group-I; 8, GLO3; 9, ACA2; 10, PAR1 group-II; 11, GLO2; 12, UNC1; 13, GLO6; 14, GIG1; 15, GLO4; 16, UNC2; 17, GLO7; 18, SCU1; 19, GIG2; 20, ACA3.