Figures & data
Table 1 Physicochemical characteristics of the soil samples
Figure 1 Bacterial community structures assessed by denaturing gradient gel electrophoresis (DGGE) analysis. (a) The DGGE banding profiles of the six soil samples and three treatments. Lanes 1–3, soil sample collected from the continuous rice field from Niigata (NC); lanes 4–6, soil sample collected from the rice–soybean rotation field from Niigata (NR); lanes 7–9, soil sample collected from the continuous rice field from Yamagata (YC); lanes 10–12, soil sample collected from the rice–soybean rotation field from Yamagata (YR); lanes 13–15, soil sample collected from the continuous rice field from Kumamoto (KC); lanes 16–18, soil sample collected from the rice–soybean rotation field from Kumamoto (KR); BA, samples without incubation; SU, samples incubated with added succinate; NS, samples incubated with added nitrate and succinate. (b) Principal component analysis (PCA) plot based on the DGGE profile. The normalized location and intensity of each DGGE band were used in the PCA analysis. The numbers in the plot correspond to the lane numbers in (a). The numbers in parentheses are the percentages of variation explained by each component.
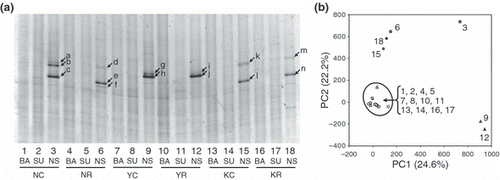
Table 2 Taxonomic assignment of the clones obtained from the excised denaturing gradient gel electrophoresis bands
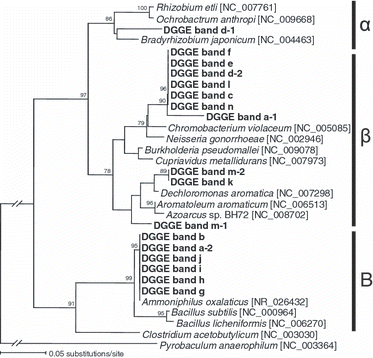
Figure 2 Neighbor-joining tree showing the phylogenetic relationship between the clones obtained from the excised denaturing gradient gel electrophoresis band shown in Fig. 1. Bootstrap values (%) were generated from 1,000 replicates and only values >70% are shown. α, Alphaproteobacteria; β, Betaproteobacteria; B, Bacilli.