Figures & data
TABLE 1 Gene disruption efficiency after zygotic injectionTable Footnotea
FIG 1 Generation and genotyping of founder mice produced by TALEN-Agouti mRNAs. (A) Schematic drawing of TALEN plasmids containing three N-terminal Flag epitopes, a nuclear localization signal (NLS), 20 TALE modules, and a fused C-terminal FokI nuclease. The positions of primers used for sequence verification of the targeting plasmids are indicated below the figure. Plasmids transfected into 293T cells encoded anti-Flag precipitable proteins of predicted masses. (B) Schematic illustration of binding of forward (black) and reverse (blue) TALEN pairs separated by 14-bp spacer sequences (red) at the Agouti locus. Host flanking sequences, including the “A” of the initiation codon (left) are shown in purple. (C) Litter of six F0 pups (two black) generated from a foster mother implanted with zygotes injected with Agouti-TALEN pairs. (D) Primers used for PCR amplification and nucleotide sequencing of tail DNAs of F0 mice; the box indicates sequences encoding TALEN binding sites and flanking sequences between primers F1 and F2. (E) Nucleotide sequences of tail DNAs from TALEN-Agouti (Ag) F0 mice; the color code is identical to that shown in panel B. Black and brown pups are indicated by the corresponding Ag font colors (left). Mutated alleles detected in each F0 mouse are summarized at the right. Symbols: WT, wild type; Δ, deletion; –, number of base pairs deleted (e.g., “−10” indicates 10 bp deleted).
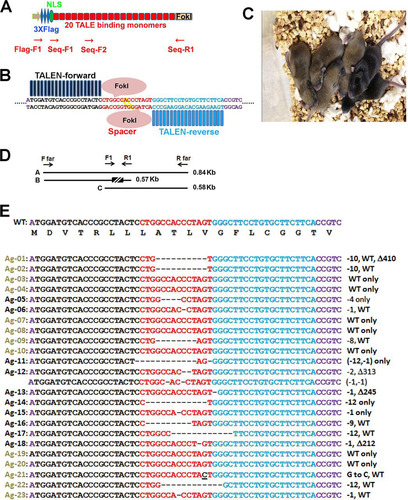
FIG 2 Agouti (Ag) genotypes of F1 pups derived from breeding seven Ag F0 mice to wild-type C57BL/6 mice. The nucleotide sequences of forward (black) and reverse (blue) TALEN binding sequences at the wild-type Agouti locus are shown at the top. Mutations detected in individual F0 mice (top) and their respective F1 offspring (bottom) are listed. Each Ag founder mouse (corresponding to those in ) with the number of derived F1 pups of indicated genotypes (parenthesis) are shown at the left. Mutated alleles summarized at the right (as for ) include additional large deletions (Δ) detected using F-far and R-far primers () and confirmed by nucleotide sequencing. Black and brown F0 mice are denoted by corresponding Ag font colors at the left. Symbols: WT, wild type; Δ, deletion; –, number of base pairs deleted (e.g., “−10” indicates 10 bp deleted).
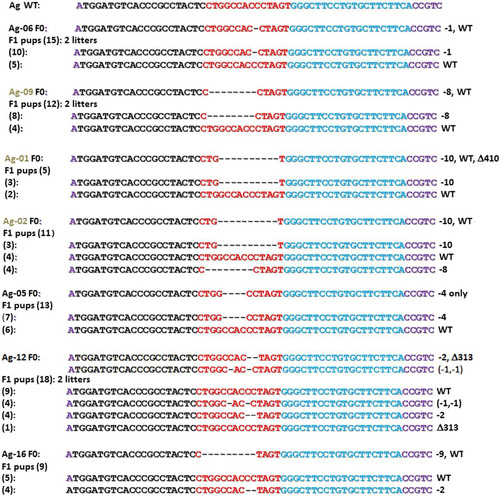
FIG 3 Nucleotide sequences of founder AgMi mice derived after zygotic coinjection of TALEN mRNA pairs directed to the Agouti (Ag) and miR-205 (Mi) loci. The tail DNAs of individually numbered AgMi mice (left margin) were genotyped for Agouti (A) and miR-205 (B) mutations, summarized at the right as for . Sequences bound by forward (black) and reverse (blue) TALENs interrupted by spacer sequences (red) are color coded. The nucleotide sequence of pre-miR-205 shown in panel B denotes one of two strands (orange box) of processed miR-205. Seed sequences are denoted by the purple bracket. Mutated alleles summarized at the right include additional large deletions (Δ). Black and brown F0 mice are denoted by corresponding AgMi font colors. Symbols: WT, wild type; Δ, deletion; –, number of base pairs deleted (e.g., “−11” indicates 11 bp deleted).
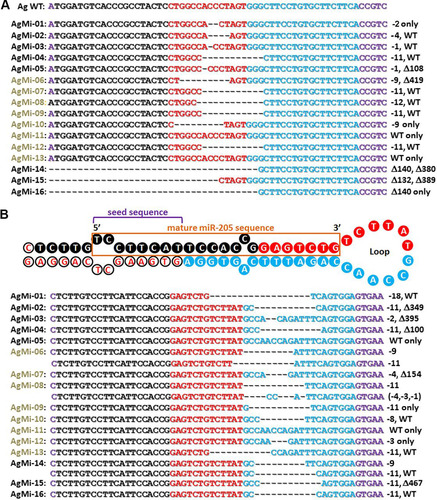
TABLE 2 Simultaneous inheritance of Agouti and miR-205 mutations in AgMi miceTable Footnotea
