Figures & data
FIG 1 The circ_0032463/miR-498/LEF1 axis might act as a key regulatory pathway in osteosarcoma progression. (A) Two upregulated circRNAs were screened out by a P value of <0.05 and a logFC of >1.5 in a GEO data set, GSE96964. (B and C) RT-qPCR analysis revealed circ_0032463 expression in osteosarcoma tissues was higher than that in normal tissues. (D) The molecular structure of circ_0032463. (E) Venn diagram showing 27 upregulated genes overlapped from GSE16088 and GSE12865 based on an adj.P value of <0.05 and logFC of >2. (F) STRING analysis demonstrates LEF1 as a key gene involving cell adhesion and cell migration among the 27 upregulated genes. (G) Venn diagram showing three miRNAs potentially interact with both circ_0032463 and LEF1. CircInteractome predicted the miRNAs binding to circ_0032463. TargetScan and starBase predicted the miRNAs binding to LEF1. (H to J) RT-qPCR analysis determined miR-498 was expressed lowest in osteosarcoma tissues among the three miRNAs.
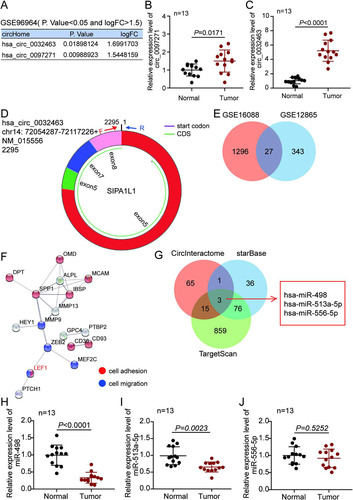
FIG 2 circ_0032463 is upregulated in osteosarcoma cells. (A) Measurement of circ_0032463 expression in osteosarcoma cell lines (U2OS, HOS, Saos-2, SOSP-9607, and SW1353) and the normal osteoblast cell line (hFOB1.19) by RT-qPCR. (B) Measurement of circ_0032463 expression in the nucleus and cytoplasm of U2OS and Saos-2 cells by RT-qPCR. (C) RT-qPCR analysis of circ_0032463 and SIPA1L1 mRNA after treatment with or without RNase R in U2OS and Saos-2 cells. *, P < 0.05; **, P < 0.001.
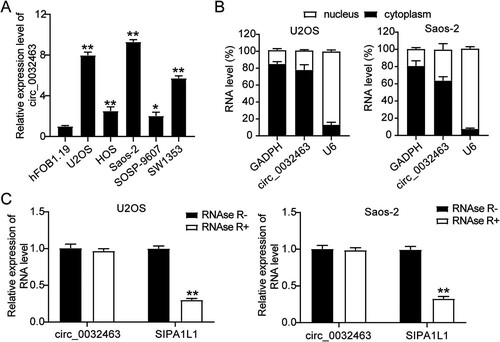
FIG 3 circ_0032463 promoted cell proliferation, adhesion, and migration but suppressed apoptosis of osteosarcoma cells. (A) RT-qPCR analysis of circ_0032463 level in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463#1, and sh-circ_0032463#2. (B) Cell viability was detected in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463#1, and sh-circ_0032463#2 at 0, 24, 48, and 72 h by CCK-8 assay. (C) Cell proliferation was detected in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463#1, and sh-circ_0032463#2 by BrdU assay. (D) Cell apoptosis was determined in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463#1, and sh-circ_0032463#2 by FITC apoptosis detection kit. (E) Cell adhesion was determined in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463#1, and sh-circ_0032463#2 at 30 and 60 min, respectively. (F) Cell migration ability was determined in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463#1, and sh-circ_0032463#2 by wound healing assay. sh-NC, shRNA-negative control; sh-circ_0032463#1 and sh-circ_0032463#2 are shRNAs of circ_0032463#1 and circ_0032463#2, respectively. *, P < 0.05; **, P < 0.001.
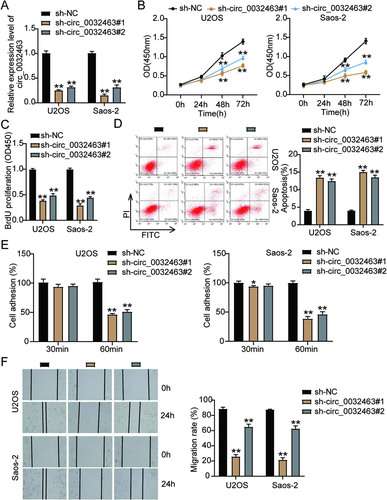
FIG 4 circ_0032463 directly interacted with miR-498 in osteosarcoma cells. (A) Measurement of miR-498 expression in osteosarcoma cell lines (U2OS, HOS, Saos-2, SOSP-9607, and SW1353) and the normal osteoblast cell line (hFOB1.19) by RT-qPCR. (B) The correlation between the relative expression level of miR-498 and circ_0032463 was evaluated by Pearson’s correlation test. (C) The potential binding sequences of miR-498 in circ_0032463 was predicted by CircInteractome. (D) RT-qPCR analysis of miR-498 expression in U2OS and Saos-2 cells transfected with NC mimics and miR-498 mimics. (E) The interaction of circ_0032463 and miR-498 was determined by RIP assay. (F) The enrichment of circ_0032463 in U2OS and Saos-2 cells transfected with NC mimics and miR-498 mimics was assessed by RIP assay. *, P < 0.05; **, P < 0.001. NC, negative control; Wt, wild type; Mut, mutant.
FIG 5 Overexpression of wild-type circ_0032463 but not circ_0032463 with mutant miR-498 binding sites promoted tumorigenesis in osteosarcoma cells. (A) Measurement of miR-498 and circ_0032463 expression in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ by RT-qPCR. (B) Cell viability was detected in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ. (C) Cell proliferation was detected in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ. (D) Cell apoptosis was detected in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ. (E) Cell adhesion was detected in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ at 30 and 60 min. (F) Cell migration was detected in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ. OE WT-circ, wild-type circ_0032463 overexpression vector; OE Mut-circ, circ_0032463 overexpression vector with mutant miR-498 binding sites. *, P < 0.05; **, P < 0.001.
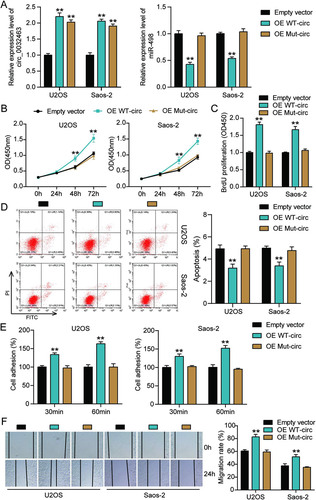
FIG 6 circ_0032463 accelerated the tumorigenesis in osteosarcoma cells by inhibiting miR-498. (A) Measurement of miR-498 and circ_0032463 expression in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-circ_0032463, miR-inhibitor, and sh-circ_0032463+miR-inhibitor by RT-qPCR. (B) Cell viability was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-circ_0032463, miR-inhibitor, and sh-circ_0032463+miR-inhibitor by CCK-8 assay. (C) Cell proliferation was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-circ_0032463, miR-inhibitor, and sh-circ_0032463+miR-inhibitor by BrdU assay. (D) Cell apoptosis was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-circ_0032463, miR-inhibitor, and sh-circ_0032463+miR-inhibitor with the FITC apoptosis detection kit. (E) Cell adhesion was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-circ_0032463, miR-inhibitor, and sh-circ_0032463+miR-inhibitor at 30 and 60 min. (F) Cell migration was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-circ_0032463, miR-inhibitor, and sh-circ_0032463+miR-inhibitor by wound healing assay. sh-NC, shRNA-negative control; NC inhibitor, miR-498 inhibitor negative control; sh-circ_0032463, shRNA-circ_0032463; sh-circ_0032463+miR-inhibitor, shRNA-circ_0032463 plus miR-498 inhibitor. *, P < 0.05; **, P < 0.001.
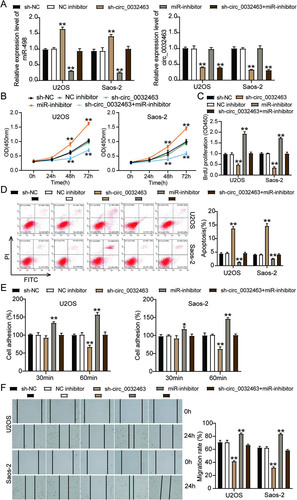
FIG 7 miR-498 was directly targeted with LEF1 in osteosarcoma cells. (A) RT-qPCR analysis of LEF1 expression in osteosarcoma tissues (n = 13) and normal tissues (n = 13). (B) Measurement of LEF1 expression in osteosarcoma cell lines (U2OS, HOS, Saos-2, SOSP-9607, and SW1353) and the normal osteoblast cell line (hFOB1.19) using RT-qPCR. (C) Pearson’s correlation analysis of the correlation between the relative expression level of miR-498 and LEF1. (D) The potential binding sequences of miR-498 on the LEF1 3′ UTR were predicted by TargetScan. (E) A dual-luciferase assay was performed to reveal the luciferase activity in cells cotransfected with plasmid LEF1 3′UTR-Wt or LEF1 3′UTR-Mut and NC mimics or miR-498 mimics. (F) The RNA pulldown assay determined the enrichment of LEF1 in U2OS and Saos-2 cells transfected with Bio-NC and Bio-miR-498. (G) RT-qPCR analysis of LEF1 expression in U2OS and Saos-2 cells transfected with sh-NC, sh-circ_0032463, and sh-circ_0032463+miR-inhibitor. (H) Measurement of LEF1 protein level in U2OS and Saos-2 cells transfected with empty vector, OE WT-circ, and OE Mut-circ by Western blot assay. NC mimics, negative-control mimics; Wt, wild type; Mut, mutant; Bio-NC, biotin-negative control; Bio-miR-498, biotin–miR-498; sh-NC, shRNA-negative control; sh-circ_0032463, shRNA-circ_0032463; miR-inhibitor, miR-498 inhibitor; OE WT-circ, wild-type circ_0032463 overexpression vector; OE Mut-circ, circ_0032463 overexpression vector with mutant miR-498 binding sites. *, P < 0.05; **, P < 0.001.
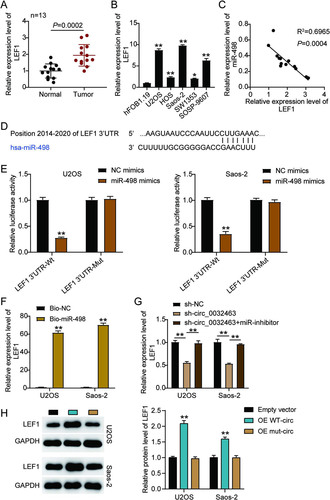
FIG 8 miR-498 attenuated tumor development by repressing LEF1 in osteosarcoma cells. (A) Measurement of miR-498 and LEF1 expression in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor by RT-qPCR. (B) Measurement of LEF1 protein expression in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor by Western blot assay. (C) Cell viability was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor by CCK-8 assay. (D) Cell proliferation was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor by BrdU assay. (E) Cell apoptosis was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor with the FITC apoptosis detection kit. (F) Cell adhesion was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor at 30 and 60 min. (G) Cell migration was detected in U2OS and Saos-2 cells transfected with sh-NC, NC inhibitor, sh-LEF1, miR-inhibitor, and sh-LEF1+miR-inhibitor by wound healing assay. sh-NC, shRNA-negative control; NC inhibitor, negative control of miR-498 inhibitor; sh-LEF1, shRNA-LEF1; miR-inhibitor, miR-498 inhibitor; sh-LEF1+miR-inhibitor, shRNA-LEF1 plus miR-498 inhibitor. *, P < 0.05; **, P < 0.001.
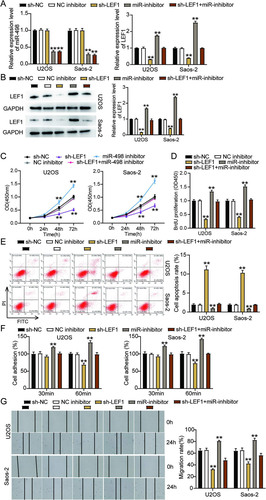
FIG 9 Overexpression of LEF1 with mutant but not wild-type miR-498 binding sites reversed the ameliorated tumorigenesis in circ_0032463-silenced osteosarcoma cells. (A) Measurement of circ_0032463 and LEF1 expression in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, or sh-circ+OE Mut-LEF1 by RT-qPCR. (B) Measurement of LEF1 protein expression in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, or sh-circ+OE Mut-LEF1 by Western blot assay. (C) Cell viability was detected in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, and sh-circ+OE Mut-LEF1. (D) Cell proliferation was detected in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, or sh-circ+OE Mut-LEF1 by BrdU assay. (E) Cell apoptosis was detected in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, or sh-circ+OE Mut-LEF1 by FITC assay. (F) Cell adhesion was detected in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, or sh-circ+OE Mut-LEF1 at 30 and 60 min. (G) Cell migration was detected in U2OS and Saos-2 cells transfected with sh-NC+Empty vector, sh-circ+Empty vector, sh-circ+OE WT-LEF1, sh-circ+OE Mut-LEF1 by wound healing assay. sh-NC+Empty vector, shRNA-negative control plus empty vector; sh-circ+Empty vector, shRNA-circ_0032463 plus empty vector; sh-circ+OE WT-LEF1, shRNA-circ_0032463 plus wild-type circ_0032463 overexpression vector; sh-circ+OE Mut-LEF1, shRNA-circ_0032463 plus circ_0032463 overexpression vector with mutant miR-498 binding sites. *, P < 0.05, and **, P < 0.001, versus the sh-NC+Empty vector group; #, P < 0.05, and ##, P < 0.001, versus the sh-circ+Empty vector group.
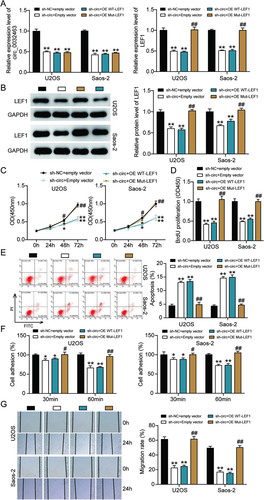
TABLE 1 Clinical characteristics of 13 osteosarcoma patients
TABLE 2 Primer sequences for qRT-PCR
