Figures & data
FIG 1 SARS-CoV-2 protein functions and host interactions. Shown is a graphic representation of the SARS-CoV-2 proteome. Boxes connected to proteins indicate the virus transcription and translation functions (italics) (summary of ) and known human protein interaction with their effect on human pathways (summary of ). Abbreviations: 3CLpro, 3-chymotrypsin-like protease; 7SL, RNA component of SRP ribonucleoprotein complex; ACE2, angiotensin I-converting enzyme 2; eIFs, eukaryotic translation initiation factors; ExoN, exoribonuclease; HOPS, homotypic fusion and protein sorting complex; IFN, interferon; IRF3, interferon regulatory factor 3; ISG, interferon-stimulated gene; KPNA2, karyopherin alpha 2; MAVS, mitochondrial antiviral-signaling protein; Mpro, main protease; NEMO, NF-κB essential modulator; NLRP12, NLR family pyrin domain containing 12; NUP98, nuclear pore complex protein 98; NXF1, nuclear RNA export factor 1; PARPs, poly(ADP-ribosyl) polymerases; PIC, ribosomal preinitiation complex; PLpro, papain-like protease; RAE1, RNA export 1; RdRp, RNA-dependent RNA polymerase; RIG-I, retinoic acid-inducible gene I; SRP, signal recognition particle; TAB1, TGF-β-activated kinase 1 (MAP3K7) binding protein 1; TBK1, TANK binding kinase 1; TMPRSS2, transmembrane serine protease 2; TOM70, translocase of outer membrane 70 (subunit of mitochondrial import receptor); VPS39, vacuolar protein sorting 39 (subunit of HOPS complex).
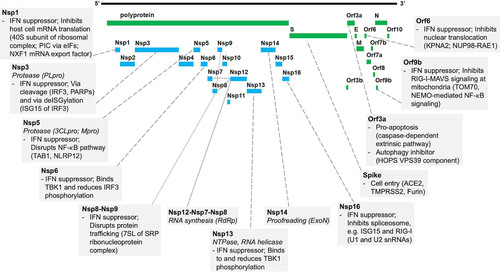
TABLE 1 SARS-CoV-2 proteins for virus transcription and translationTable Footnotea
TABLE 2 SARS-CoV-2 proteins interacting with intracellular host pathwaysTable Footnotea
TABLE 3 Inhibitors of SARS-CoV-2 transcription and translation
TABLE 4 Inhibitors of SARS-CoV-2 host protein-protein interactions
