Figures & data
FIG 1 A p62 splicing variant lacking the Keap1-interacting region is present in mice. (A) Schematic diagram of genome structures of mouse p62. Coding exons, numbered in accordance with the initiation site (exon 1), are depicted by white boxes. The full-length mRNA and its splicing variant produced from the p62 allele are shown. wt, wild type. (B) Domain structures of mouse full-length and variant p62 proteins. Phox and Bem1 (PB1) mediates homo-oligomerization or hetero-oligomerization. The LC3-interacting region (LIR) and the C-terminal ubiquitin-associated (UBA) domain promote the sequestration of ubiquitinated substrates into the autophagosome. The Keap1-interacting region (KIR) binds to Keap1. ZZ, zinc finger; TB, TRAF6-binding domain; NLS, nuclear localization signal; NES, nuclear export signal. (C) Alignment of regions containing the LIR, the KIR, and the UBA domain in the full-length and variant p62 proteins of mice. (D) RT-PCR. Analyses of p62 exon 7 splicing in MEFs, Hepa-1 cells, and mouse liver were performed. Diagrams at left indicate cDNAs encoding full-length and variant p62 proteins. The positions of primers used for RT-PCR analysis of p62 cDNA and the expected sizes of the PCR products in the presence and absence of the last half of exon 7 are shown. The right panel shows the results of gel electrophoresis (2% agarose gel) of RT-PCR products from cDNAs of MEFs, Hepa-1 cells, and mouse liver. (E) Digital PCR analysis. RNA copy numbers for full-length p62 and its variant in MEFs, Hepa-1 cells, and mouse liver were determined using a QuantStudio 3D digital PCR system. (F) Immunoblot analysis. Total cell lysates of MEFs and Hepa-1 cells and mouse liver homogenate were prepared and subjected to immunoblot analysis with anti-p62 antibody. Nontagged full-length or variant p62 was expressed in p62-deficient HeLa cells, and the lysates were used as positive controls. Data are representative of three independent experiments. Quantitative densitometry analysis of immunoblotting data was performed, and the levels of full-length p62 and its variant were normalized against that of actin. (G) Immunoblot analysis. Primary mouse hepatocytes were challenged with 10 μM sodium arsenite [As(III)] for the indicated times. Data are representative of three independent experiments. Quantitative densitometry analysis of immunoblotting data was performed, and the levels of full-length p62 and its variant were normalized against that of actin.
![FIG 1 A p62 splicing variant lacking the Keap1-interacting region is present in mice. (A) Schematic diagram of genome structures of mouse p62. Coding exons, numbered in accordance with the initiation site (exon 1), are depicted by white boxes. The full-length mRNA and its splicing variant produced from the p62 allele are shown. wt, wild type. (B) Domain structures of mouse full-length and variant p62 proteins. Phox and Bem1 (PB1) mediates homo-oligomerization or hetero-oligomerization. The LC3-interacting region (LIR) and the C-terminal ubiquitin-associated (UBA) domain promote the sequestration of ubiquitinated substrates into the autophagosome. The Keap1-interacting region (KIR) binds to Keap1. ZZ, zinc finger; TB, TRAF6-binding domain; NLS, nuclear localization signal; NES, nuclear export signal. (C) Alignment of regions containing the LIR, the KIR, and the UBA domain in the full-length and variant p62 proteins of mice. (D) RT-PCR. Analyses of p62 exon 7 splicing in MEFs, Hepa-1 cells, and mouse liver were performed. Diagrams at left indicate cDNAs encoding full-length and variant p62 proteins. The positions of primers used for RT-PCR analysis of p62 cDNA and the expected sizes of the PCR products in the presence and absence of the last half of exon 7 are shown. The right panel shows the results of gel electrophoresis (2% agarose gel) of RT-PCR products from cDNAs of MEFs, Hepa-1 cells, and mouse liver. (E) Digital PCR analysis. RNA copy numbers for full-length p62 and its variant in MEFs, Hepa-1 cells, and mouse liver were determined using a QuantStudio 3D digital PCR system. (F) Immunoblot analysis. Total cell lysates of MEFs and Hepa-1 cells and mouse liver homogenate were prepared and subjected to immunoblot analysis with anti-p62 antibody. Nontagged full-length or variant p62 was expressed in p62-deficient HeLa cells, and the lysates were used as positive controls. Data are representative of three independent experiments. Quantitative densitometry analysis of immunoblotting data was performed, and the levels of full-length p62 and its variant were normalized against that of actin. (G) Immunoblot analysis. Primary mouse hepatocytes were challenged with 10 μM sodium arsenite [As(III)] for the indicated times. Data are representative of three independent experiments. Quantitative densitometry analysis of immunoblotting data was performed, and the levels of full-length p62 and its variant were normalized against that of actin.](/cms/asset/aa7df85d-f7ad-405e-bf72-4ae09ec6ea62/tmcb_a_12276738_f0001.jpg)
FIG 2 The p62 splicing variant forms a complex with full-length p62. (A) Gel filtration chromatography. FLAG-tagged p62 or its variant was expressed in p62-knockout MEFs. Lysates were subjected to gel filtration chromatography followed by immunoblot analysis with anti-p62 antibody. Data are representative of three independent experiments. (B) Gel filtration chromatography. FLAG-tagged PB1 mutant p62 or its variant form was expressed in p62-knockout MEFs. Lysates were subjected to gel filtration chromatography followed by immunoblot analysis with anti-p62 antibody. Data are representative of three independent experiments. (C) Gel filtration chromatography. FLAG-tagged full-length p62 and GFP-tagged variant p62 were coexpressed in p62−/− MEFs, and cell lysates were subjected to gel filtration chromatography followed by immunoblot assay with anti-p62 antibody. Data are representative of three independent experiments. (D) Immunoprecipitation (IP) assay. Fractions 13 to 15 from the gel filtration chromatography shown in panel B were subjected to immunoprecipitation with anti-FLAG or anti-GFP antibody. The immunoprecipitates were examined by immunoblotting with anti-p62 antibody. Data are representative of three independent experiments.
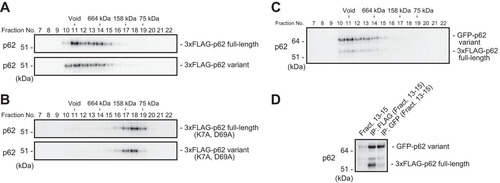
FIG 3 The p62 variant lacks the ability to bind Keap1. (A) Immunoprecipitation assay. FLAG-tagged full-length or variant p62 was expressed in p62−/− MEFs, and cell lysates were subjected to immunoprecipitation with anti-FLAG antibody. The immunoprecipitates were examined by immunoblotting with the indicated antibodies. Data are representative of three independent experiments. (B) Immunofluorescence analysis. 3×FLAG-tagged p62 or its variant was expressed in p62-knockout Huh1 cells by use of an adenovirus vector system, and the cells were immunostained with anti-FLAG and anti-Keap1 antibodies. Each inset is a magnified image. Bar, 20 μm.
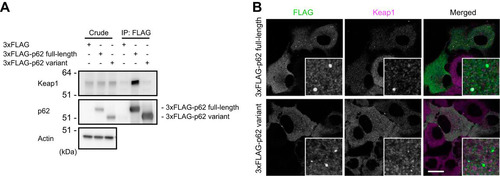
FIG 4 The p62 variant is degraded by autophagy. (A) Immunoprecipitation assay. One-Strep-FLAG (OSF)-tagged LC3B, GABARAPL2, and the corresponding hydrophobic pocket mutants were coexpressed with GFP-tagged full-length or variant p62 in HEK293T cells. Precipitates generated with anti-FLAG antibody were subjected to immunoblot analysis with anti-GFP and anti-FLAG antibodies. Data are representative of three independent experiments. (B) Immunoblot analysis. Primary hepatocytes were cultured in the presence or absence of E64d and pepstatin A (Pep. A) for 24 h. The lysates were subjected to NuPAGE followed by immunoblot analysis with the indicated antibodies. Data are representative of three independent experiments. (C) Immunoblot analysis. Homogenates prepared from livers of control Atg7f/f and Atg7f/f; albumin-Cre mice were analyzed by immunoblotting with the indicated antibodies. Data are representative of three independent experiments.
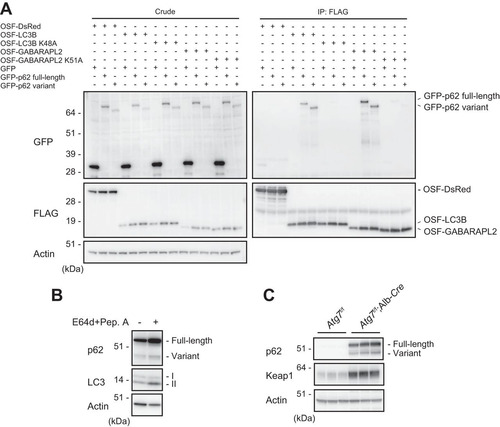
FIG 5 The p62 variant increases the amount of Keap1 and represses Nrf2 activity. (A) Real-time PCR. Relative mRNA levels of Nrf2 targets in primary mouse hepatocytes expressing full-length p62 or variant p62 at the indicated multiplicities of infection (MOIs) are shown. Values were normalized to the amount of each mRNA in the nontreated wild-type hepatocytes. The experiments were performed three times. Data are means ± standard errors of the means (SEM). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (as determined by Welch's t test). (B) Immunoblot analysis. Cytosolic and nuclear fractions were prepared from the hepatocytes prepared as described for panel A and subjected to immunoblot analysis with the indicated antibodies. Data are representative of three independent experiments. (C) Ubiquitination assay with Nrf2. FLAG-tagged p62 and its variant were overproduced in primary mouse hepatocytes. Cells were cultured in the absence or presence of 10 μM lactacystin for 4 h. The cell lysates were immunoprecipitated with anti-Nrf2 antibody, subjected to electrophoresis in a NuPAGE gel, and analyzed by immunoblotting with anti-Nrf2 and antiubiquitin antibodies. Data are derived from three separate experiments. (D) Immunoblot analysis. Total lysates were prepared from primary mouse hepatocytes expressing full-length p62 or its variant, alone or in combination, at the indicated MOIs and then subjected to immunoblot analysis. Data are representative of three independent experiments. (E) Immunoblot analysis. Total lysates were prepared from p62-deficient primary mouse hepatocytes expressing full-length p62 alone or together with the variant at the indicated MOIs. The lysates were subjected to immunoblot analysis. Data are representative of three independent experiments. (F) Real-time PCR. Relative mRNA levels of Nrf2 targets in hepatocytes prepared as described for panel E are shown. Values were normalized to the amount of mRNA in nontreated p62-deficient hepatocytes. The experiments were performed three times. Data are means ± SEM. *, P < 0.05; **, P < 0.01 (as determined by Welch's t test).
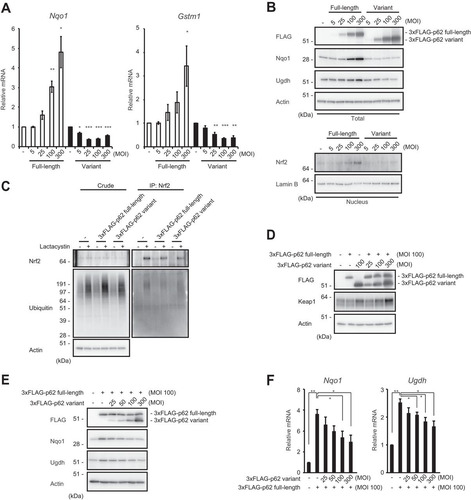
FIG 6 The p62 variant negatively regulates the p62-Keap1-Nrf2 axis in mouse hepatocytes. (A) Schematic diagram of the genome structures of wild-type p62 and p62-GFP knock-in alleles. Coding exons, numbered in accordance with the initiation site (exon 1), are depicted by white boxes. E2-cDNA–GFP–pA indicates the p62 cDNA fragment (positions 302 to 1326) fused with GFP cDNA and simian virus 40 (SV40) poly(A). mRNAs and proteins produced from each allele are shown. (B) Immunoblot analysis. Primary hepatocytes prepared from both wild-type and p62-GFP knock-in mice were challenged with 10 μM sodium arsenite [As(III)] for 12 h. After removal of As(III), cells were cultured in regular medium for the indicated times. Cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies. Data show the results of two independent experiments. Numerical values indicate the results of quantitative densitometric analyses of Nqo1 normalized against the levels in nontreated cells. An asterisk indicates a nonspecific band. (C) Real-time PCR. Relative mRNA levels of Nrf2 targets in hepatocytes prepared as described for panel B are shown. Values were normalized against the corresponding mRNA levels in nontreated wild-type or p62-GFP knock-in hepatocytes. The experiments were performed two times.
![FIG 6 The p62 variant negatively regulates the p62-Keap1-Nrf2 axis in mouse hepatocytes. (A) Schematic diagram of the genome structures of wild-type p62 and p62-GFP knock-in alleles. Coding exons, numbered in accordance with the initiation site (exon 1), are depicted by white boxes. E2-cDNA–GFP–pA indicates the p62 cDNA fragment (positions 302 to 1326) fused with GFP cDNA and simian virus 40 (SV40) poly(A). mRNAs and proteins produced from each allele are shown. (B) Immunoblot analysis. Primary hepatocytes prepared from both wild-type and p62-GFP knock-in mice were challenged with 10 μM sodium arsenite [As(III)] for 12 h. After removal of As(III), cells were cultured in regular medium for the indicated times. Cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies. Data show the results of two independent experiments. Numerical values indicate the results of quantitative densitometric analyses of Nqo1 normalized against the levels in nontreated cells. An asterisk indicates a nonspecific band. (C) Real-time PCR. Relative mRNA levels of Nrf2 targets in hepatocytes prepared as described for panel B are shown. Values were normalized against the corresponding mRNA levels in nontreated wild-type or p62-GFP knock-in hepatocytes. The experiments were performed two times.](/cms/asset/6d8e7641-5a9b-4a17-94fb-c969aff3913d/tmcb_a_12276738_f0006.jpg)
