Figures & data
FIG 1 Cki3 is required for NETO inhibition when DNA replication is blocked. (A) Distribution of CRIB-GFP signals in pol1 and pol1 cki3Δ mutants. Cells were grown at 27°C, shifted up to 36°C, and kept at that temperature for 3.5 h. Representative patterns of CRIB-GFP (top) and the percentages of monopolar and bipolar cells at 27°C and 36°C (n > 100) (bottom) are shown. One cell undergoing monopolar growth is outlined (top right). (B) Increase in Cki3 kinase activity upon S-phase arrest. Cells expressing Cki3-GFP (the wild type [WT] and the pol1 mutant [lanes 2 and 3, respectively]) or nontagged wild-type cells (lane 1) were grown at 25°C and then transferred to 36°C and kept at that temperature for 1 h. Cki3-GFP was immunoprecipitated (IP) with an anti-GFP antibody (using 4 mg of protein extracts). Half of each precipitate was used for immunoblotting, while the other half was assayed for the phosphorylation reaction using casein as a substrate. The kinase activity (KA) was quantified with Image J (bottom). WCE, whole-cell extracts; RI, relative intensity.
![FIG 1 Cki3 is required for NETO inhibition when DNA replication is blocked. (A) Distribution of CRIB-GFP signals in pol1 and pol1 cki3Δ mutants. Cells were grown at 27°C, shifted up to 36°C, and kept at that temperature for 3.5 h. Representative patterns of CRIB-GFP (top) and the percentages of monopolar and bipolar cells at 27°C and 36°C (n > 100) (bottom) are shown. One cell undergoing monopolar growth is outlined (top right). (B) Increase in Cki3 kinase activity upon S-phase arrest. Cells expressing Cki3-GFP (the wild type [WT] and the pol1 mutant [lanes 2 and 3, respectively]) or nontagged wild-type cells (lane 1) were grown at 25°C and then transferred to 36°C and kept at that temperature for 1 h. Cki3-GFP was immunoprecipitated (IP) with an anti-GFP antibody (using 4 mg of protein extracts). Half of each precipitate was used for immunoblotting, while the other half was assayed for the phosphorylation reaction using casein as a substrate. The kinase activity (KA) was quantified with Image J (bottom). WCE, whole-cell extracts; RI, relative intensity.](/cms/asset/287c3e39-e149-4b25-a2df-30c0f7ba8e98/tmcb_a_12275595_f0001_oc.jpg)
FIG 2 Cki3 acts downstream of Cds1 and calcineurin when the DNA replication checkpoint is activated. (A) Growth patterns upon overexpression of various genes in individual deletion strains. Cells containing an empty vector (pREP1) or plasmid pREP1-cki3+, pREP1-cds1+, or pREP1-ppb1ΔC were cultured in the absence of thiamine (derepressed) at 28°C for 16 h, and growth polarity was examined by calcofluor white staining. The percentages of monopolar cells are shown. P values were obtained by chi-square tests. **, P < 0.01. (B) Percentages of monopolar, bipolar, and septated cells. Individual strains were grown at 25°C and shifted to 36°C for 4 h. The growth patterns were examined by calcofluor white staining (n > 200). (C) HU sensitivity. Tenfold serial dilutions of the indicated strains were spotted on Edinburgh minimal medium (EMM) plates containing appropriate supplements (leucine, uracil, histidine, adenine, and lysine) in the presence or absence of HU (5 and 7.5 mM) and incubated at 27°C for 4 days.
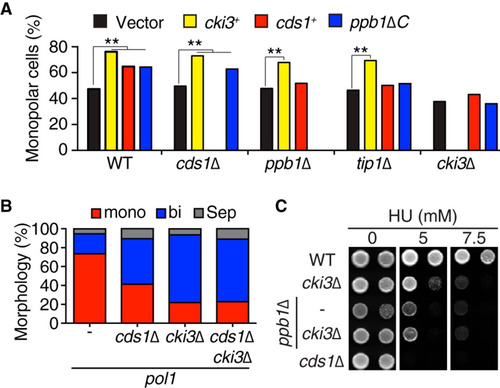
FIG 3 Cki3 acts in concert with Tip1 to retain cell viability under an S-phase block. (A) cki3Δ and tip1I87AI137A render the pol1 mutant more sensitive to high temperatures. Tenfold serial dilutions of the indicated strains were spotted on rich YE5S (yeast extract with 5 supplements) plates and incubated at various temperatures; the plates were incubated for 2 days. (B) Cell viability. (Left) The indicated strains were incubated at 36°C, and the viable colonies were counted by plating aliquots from cell cultures at each time point. (Right) Percentages of lethal “cut” cells (n > 200). (C) Functional hierarchy among individual factors required for NETO delay under conditions of perturbed DNA replication. Cki3 and Tip1 act downstream of Cds1 and calcineurin, but it is possible that the two proteins functionally interact (depicted as a two-headed arrow).
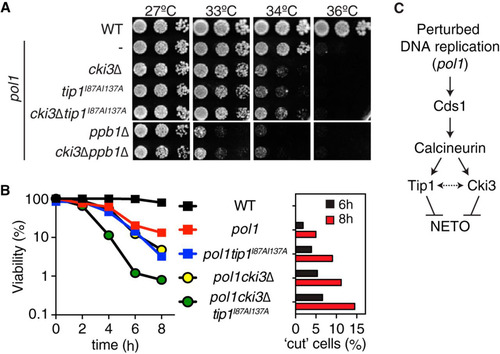
FIG 4 Cki3 is negatively regulated by autophosphorylation. (A) Phosphatase treatment of Cki3. Individual cells were grown at 25°C, and whole-cell extracts were prepared from cells expressing Cki3-HA and treated with λ-PPase in the presence or absence of the phosphatase inhibitor. Proteins were separated by SDS-PAGE in the presence (top) or absence (bottom) of 50 μM Phos-tag. Immunoblotting was performed with an anti-HA antibody. (B) The C terminus of Cki3 is autophosphorylated. Individual cells were grown at 25°C. Whole-cell extracts were prepared from strains containing the indicated Cki3 mutants tagged with HA, treated with λ-PPase, and separated by SDS-PAGE as for panel A. (C) Schematic presentation of the Cki3 structure and the amino acid sequence of the C-terminal region. The 4 amino acid residues shown in red (S and T) were mutated to either alanines (Cki34A) or glutamates (Cki34E). (D) Phosphomimetic Cki3 behaves like cki3 deletion. Cells containing CRIB-GFP were grown at 27°C, shifted up to 36°C, and kept at that temperature for 4 h. Growth polarity was examined by CRIB-GFP distribution (n > 100). Representative images (top) and quantification (bottom) are shown.
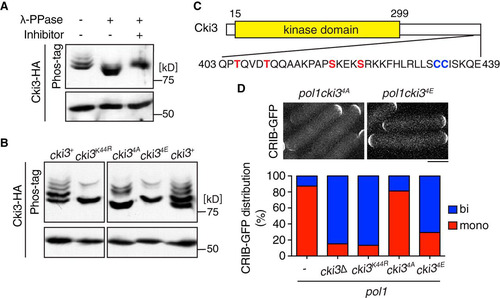
FIG 5 Calcineurin is responsible for Cki3 dephosphorylation and promotes its kinase activity. (A) Calcineurin binds Cki3. Individual cells were grown at 27°C, and whole-cell extracts were prepared from cells containing Ppb1-FLAG and Cki3-GFP or Ppb1-FLAG or Cki3-GFP only. Immunoprecipitation was performed with an anti-GFP antibody, followed by immunoblotting with anti-FLAG and anti-GFP antibodies. (B) Calcineurin is responsible for dephosphorylation of Cki3. Immunoprecipitates prepared from the indicated strains containing Cki3-HA were immunoblotted with an anti-HA antibody in a Phos-tag (top) or normal (bottom) gel. As in , Cki3 was resolved into four major bands (1 to 4). Density scan patterns are shown on the right; black, wild type; gray, dis2Δ; orange, cnb1Δ; red, ppb1Δ. A.U., arbitrary units. (C) Cki3 kinase activity is reduced in the absence of calcineurin. The in vitro kinase activity of a ppb1Δ strain containing Cki3-GFP was measured as for by using casein as a substrate (far right). Strains containing nontagged Cki3, Cki3-GFP, or Cki3K44R-GFP were also used. The kinase activity (KA) was quantified with Image J (bottom).
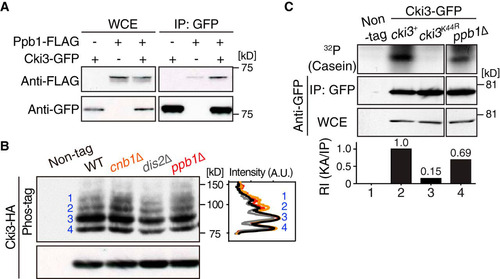
FIG 6 Erf2-mediated palmitoylation is essential for plasma membrane localization of Cki3. (A) Cki3 localization in wild-type cells. Cells exponentially growing at 27°C that produced Cki3-GFP and Crn1-tdTomato (coronin, an actin-binding protein) (Citation53) were mounted on 2% agar pads containing minimal medium. Representative images corresponding to various cell cycle stages are shown. Maximum-intensity projection images are presented. Bar, 5 μm. (B) Summary of putative palmitoyltransferases and Cki3 localization in their deletion mutants. N.D., not determined; PM, plasma membrane. (C) Cki3 localization in erf2Δ cells. Images corresponding to a single section in the middle (left) and maximum-intensity projections (right) are shown for wild-type (WT) and erf2Δ cells. Bar, 5 μm.
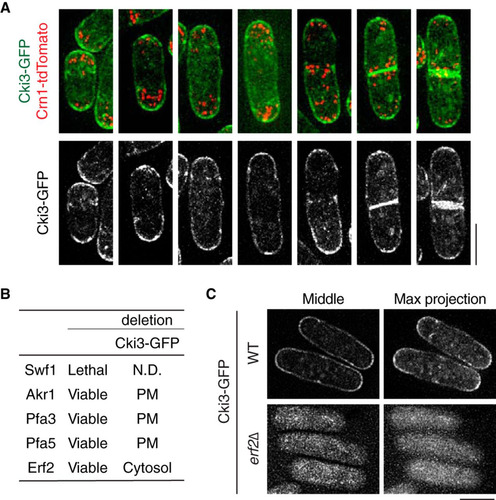
FIG 7 The C-terminal Cys-Cys sequences of Cki3 are required for not only membrane localization but also NETO delay. (A) C-terminal amino acid sequences of various CK1γ family members: S. pombe (S.p), Saccharomyces cerevisiae (S.c), C. elegans (C.e), Xenopus laevis (X.l), Drosophila melanogaster (D.m), and Homo sapiens (H.s). (B) Localization of Cki3 mutants defective in palmitoylation (Cki3SS) and kinase activity (Cki3K44R). Two cysteines were replaced with serines in Cki3SS, whereas K44, a crucial lysine residue for protein kinase activity (Citation25), was replaced with arginine in Cki3K44R. The top row shows maximum-intensity projection images, while the bottom row displays single-section (middle) images. Bar, 10 μm. (C) Reduced in vitro kinase activity in palmitoylation-deficient Cki3SS. Cell extracts (4 mg) were prepared from the indicated strains exponentially growing at 25°C, and immunoprecipitation was performed with anti-HA antibodies. Half of each precipitate was immunoblotted with an anti-GFP antibody, while the other half was assayed for the phosphorylation reaction using casein as a substrate. The kinase activity (KA) was quantified with Image J (bottom). (D) CRIB-GFP distribution. The indicated strains were grown at 27°C, shifted up to 36°C, and kept at that temperature for 3.5 h. Quantification of CRIB-GFP localization patterns is shown at the bottom (n > 100). Bar, 10 μm.
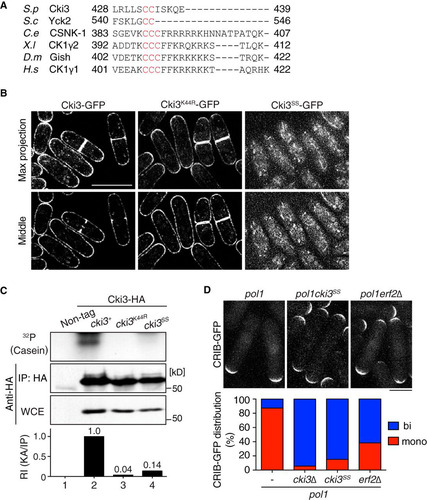
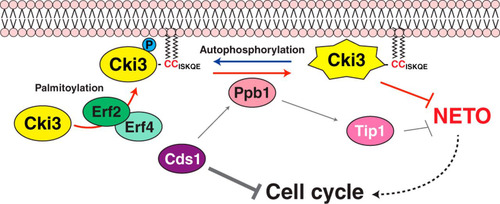
FIG 8 Schematic model of NETO controlled by Cki3. Upon a DNA replication block, the Cds1/Chk2 checkpoint kinase is activated, resulting in two inhibitory outcomes: one is cell cycle arrest, and the other is inhibition of NETO (transition from monopolar to bipolar growth). It was previously shown that calcineurin (Ppb1) acts downstream of Cds1, thereby dephosphorylating Tip1/CLIP170 and in turn restraining NETO (Citation11). In the current study, we show that, in addition to Tip1, membrane-bound Cki3 is also dephosphorylated, mediated by calcineurin. Cki3 is autophosphorylated, which inactivates Cki3 kinase activity. Therefore, calcineurin activates Cki3. Note that Cki3 and Tip1 may functionally interact, instead of acting independently. The plasma membrane localization of Cki3 is achieved by the Erf2-Erf4 palmitoyltransferase, which is also essential for Cki3 kinase activity. Dephosphorylated Cki3 inhibits NETO, the molecular mechanism of which is currently unknown. It is possible that membrane-bound substrate proteins required for NETO are inactivated through Cki3-mediated phosphorylation. Alternatively, substrate proteins that are inhibitory for NETO onset are activated instead.