Figures & data
Table 1. Heritability of psychiatric disorders and summary of relevant genetic studies.
Figure 1. The complex interaction of genes, environmental factors and intermediate phenotypes in biomarker research to predict different endophenotypes and outcome variables. A given gene set represents a genetic pathway that influences a specific biological process. Arcs between gene sets represent possible interactions (gene × gene, epistasis). Arrows originated from gene sets depict genetic effects on intermediate phenotypes. Intermediate phenotypes, which denote genetically determined neurobiological processes with causal role in the disease pathway, could be identified as biomarkers themselves but usually difficult to measure directly. However, they could be characterised by using different selected more easily measurable biomarkers (boxes in the middle). We suggest that those specific biomarkers are important and useful for personalised treatment in neuropsychiatric diseases that are informative for intermediate phenotypes influenced by environmental factors and could be used in the clinical diagnosis, outcome, therapeutic and/or side effect of drugs. Arcs between biomarkers represent possible combination of different biomarkers that provide relevant information to predict diagnosis, outcome or drug response/side effects. The environmental factors may cause epigenetic changes (e.g., up- or down-regulation in the expression of particular genes) or direct alterations in the measurable biomarker (e.g., exposure to a neurotoxin or brain trauma that disables or disconnects functional brain areas).
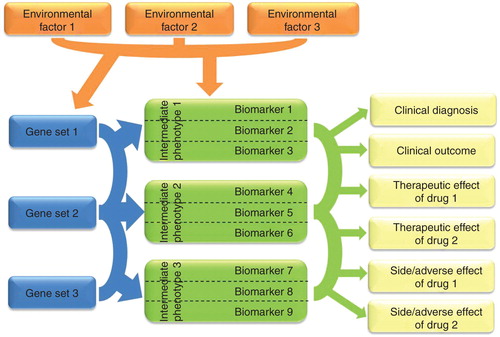
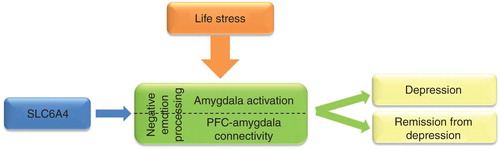
Figure 2. An example of usefulness of brain imaging intermediate phenotypes to identify risk gene and biomarker to treatment response for depression. Life stresses (e.g., watching fearful or sad faces) activate the amygdalae in the brain. The presence of a risk allele within the serotonin transporter gene (SLC6A4, 5-HTTLPR short allele) is associated with increased amygdala activation and with altered connectivity between the amygdalae and prefrontal cortical (PFC) areas compared to non-risk allele carriers during stressful situations. These biomarkers (amygdala activity and PFC–amygdala connectivity), which could be reliably measured by fMRI using selected neuropsychological tasks, provide information how the brain processes negative emotions. Impaired negative emotion processing is an intermediate phenotype that increases the risk of depression and predicts poorer antidepressant response.