Figures & data
Figure 1. CYP19A1 structure. Open boxes represent aromatase exon Is and L-shaped arrows in front of the boxes represent corresponding promoters. Closed boxes represent aromatase exons II–X encoding the open reading frame. Broken lines represent the splicing patterns.
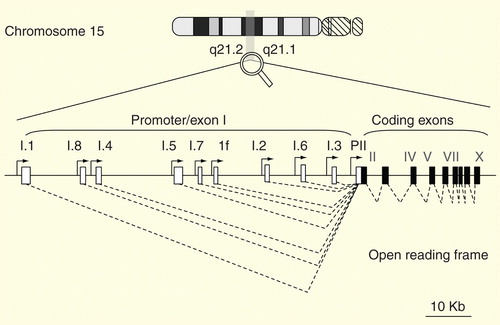
Figure 2. Clinical features of 30 male patients with molecularly diagnosed aromatase excess syndrome. (A) Distribution of gynecomastia onset. (B) Distribution of developmental stage of gynecomastia at the time of the initial visit. Severity of gynecomastia is expressed using the Tanner staging system for morphological description of the female breast. (C) Chronological change in height. Height expressed in standard deviation for age is plotted against chronological age. Closed circles, deletion-type mutations; open circles, duplication-type mutations; open triangles, inversion-type mutations. (D) Acceleration of bone growth. Differences between bone age and chronological age (years) are plotted against chronological age. Acceleration of bone growth becomes evident before 10 years of age.
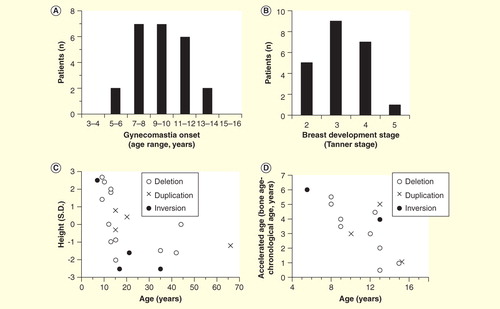
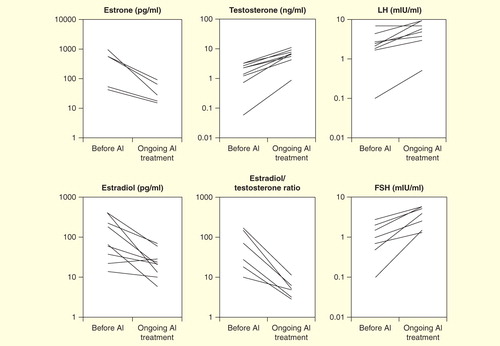
Figure 3. Hormonal profiles of 30 male patients with molecularly diagnosed aromatase excess syndrome. Isolated marks represent basal luteinizing hormone, follicle-stimulating hormone, estradiol, and testosterone levels and estradiol/testosterone ratio in each individual. Closed circles, deletion-type mutations; open circles, duplication-type mutations; open triangles, inversion-type mutations. The gray zones represent approximate normal reference ranges of adolescents. Paired marks tethered by a solid bar represent the gonadotropin levels before and 30 min after 100 µg of LH-releasing hormone loading. Paired marks in the testosterone chart represent levels before and after injection of human chorionic gonadotropin.
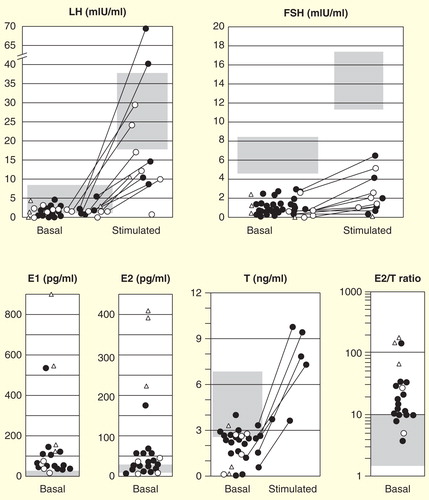
Figure 4. Schematic representations of DNA recombination events that give rise to gain-of-function mutations in familial aromatase excess syndrome. CYP19A1 exons II–X are expressed in one box. The arrows and following closed boxes represent one of multiple CYP19A1 promoters. The arrows indicate transcriptional direction. Similarly, the structure of DMXL2, a neighboring gene of CYP19A1 on the same (minus) strand as CYP19A1, is represented by a gray box (coding exons) and a shaded box (exon 1) associated with an arrow (promoter). DMXL2 exon 1 contains the 5′ end of the open reading frame. TMOD3 and SEMAD6 represent upstream and downstream neighboring genes of CYP19A1, on the opposite strand of CYP19A1, and are involved in inversion mutation. Inversion mutation may be combined with any type of other recombinations listed above to form the more complex DNA recombinations. For details, please refer to Citation[33].
![Figure 4. Schematic representations of DNA recombination events that give rise to gain-of-function mutations in familial aromatase excess syndrome. CYP19A1 exons II–X are expressed in one box. The arrows and following closed boxes represent one of multiple CYP19A1 promoters. The arrows indicate transcriptional direction. Similarly, the structure of DMXL2, a neighboring gene of CYP19A1 on the same (minus) strand as CYP19A1, is represented by a gray box (coding exons) and a shaded box (exon 1) associated with an arrow (promoter). DMXL2 exon 1 contains the 5′ end of the open reading frame. TMOD3 and SEMAD6 represent upstream and downstream neighboring genes of CYP19A1, on the opposite strand of CYP19A1, and are involved in inversion mutation. Inversion mutation may be combined with any type of other recombinations listed above to form the more complex DNA recombinations. For details, please refer to Citation[33].](/cms/asset/f526da24-1490-4a41-a56c-b2f3dff8ac31/iere_a_926810_f0004_b.jpg)
Figure 5. A diagnostic and therapeutic schema. Physical examination for estrogen-unrelated symptoms is crucial for exclusion of aromatase excess syndrome diagnosis.
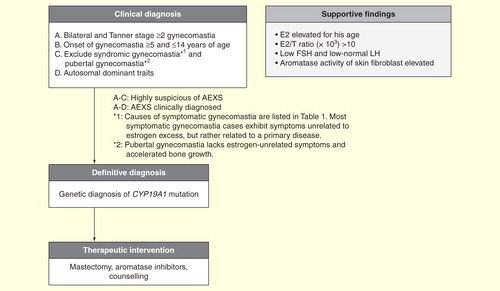
Table 1. Diseases and conditions to be distinguished from aromatase excess syndrome.