Figures & data
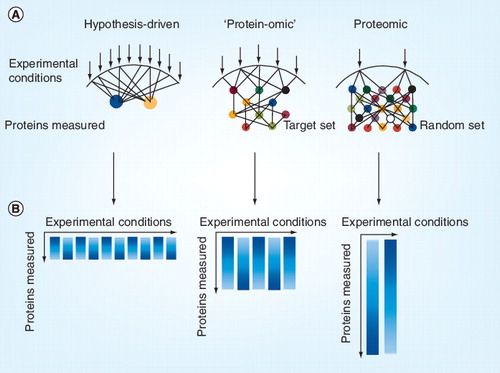
(A) Each arrow indicates an experimental condition, which may represent the stimulation or inhibition of components of a biological system. Circles represent a protein modification measured. (B) Resulting data matrix from the experimental set up. Each column represents an experimental condition and the length of the column indicates the depth of measured protein data space.
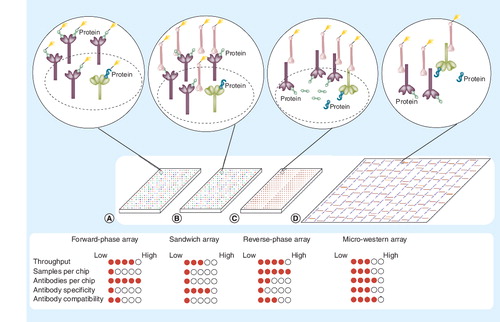
Four methodologies are indicated: (A) forward-phase arrays, where samples are labeled and applied to immobilized antibodies specific to certain proteins on nitrocellulose slides; (B) sandwich arrays, where captured proteins are detected by a second labeled antibody targeted to that protein; (C) reverse-phase lysate arrays, where complex mixtures of proteins are printed and immobilized onto slides and then specific proteins detected with a primary antibody; and (D) micro-western arrays, where samples are printed, electrophoresed to separate proteins based on size, then transferred to nitrocellulose, and then specific proteins detected with primary antibodies. A near-infrared dye-labeled secondary antibody can be used to generate fluorescent signal in (C & D) when used with the LI-COR Odyssey near-infrared scanner. Green shapes denote the targeted protein of interest while blue shapes denote proteins that nonspecifically cross-react with antibodies. The filled circles underneath each array represent the strength of each platform with respect to each criterion on the left.