Figures & data
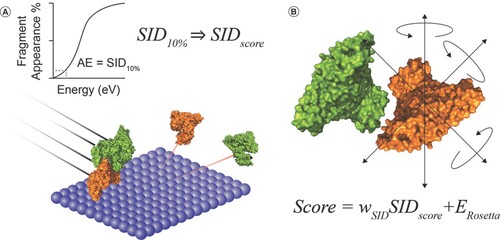
(A) A depiction of surface-induced dissociation (SID) and a graphical representation of the generation of SID appearance energy. (B) A molecule model generated in Rosetta and the Scoring function equation.
Register now or learn more
Open access
(A) A depiction of surface-induced dissociation (SID) and a graphical representation of the generation of SID appearance energy. (B) A molecule model generated in Rosetta and the Scoring function equation.

People also read lists articles that other readers of this article have read.
Recommended articles lists articles that we recommend and is powered by our AI driven recommendation engine.
Cited by lists all citing articles based on Crossref citations.
Articles with the Crossref icon will open in a new tab.