Figures & data
Table 1. In silico prediction tools of T and B-cell epitopes.
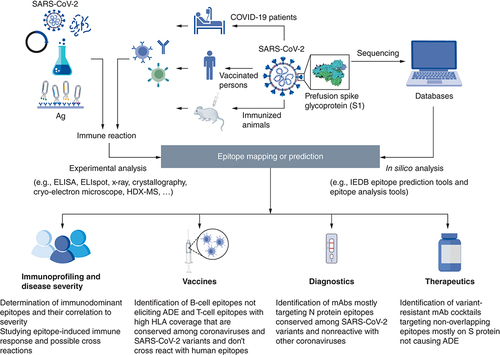
Both in silico epitope prediction tools and experimental epitope mapping are used for identification of SARS-CoV-2 epitopes. Epitope prediction and analysis tools are fed by protein sequences available in protein databases for prediction of potential epitopes that are then filtered for selected features. For experimental epitope mapping, SARS-CoV-2-specific antibodies, B-cells or T-cells may be isolated from COVID-19 patients, vaccinated persons or immunized animals. The interaction with whole, fragmented, synthetic, or recombinant antigens as well as phage display libraries can be analyzed using several experimental methods such as: ELISA, ELISpot, Cryo-electron microscope, etc.
ADE: Antibody-dependent enhancement.
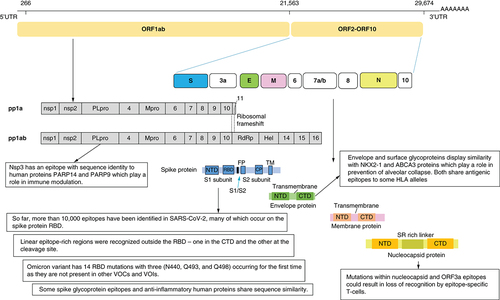
Epitope mining has revealed the presence of several immunodominant epitopes on the RBD as well as other regions of the Spike protein. Mutations in these epitopes have resulted in new variants of SARS-COV-2. Mimicry of some viral epitopes with other human proteins could provide an explanation for hyperinflammatory response.
CTD: C-terminal domain; NTD: N-terminal domain; RBD: Receptor binding domain.
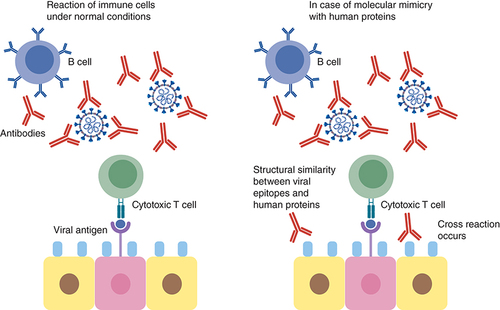
This can lead to cross reaction with the immune cells resulting in hyperinflammatory response, cytokine storm and respiratory failure.