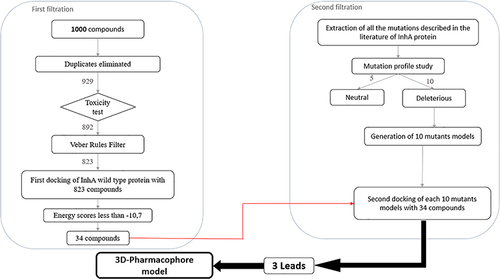
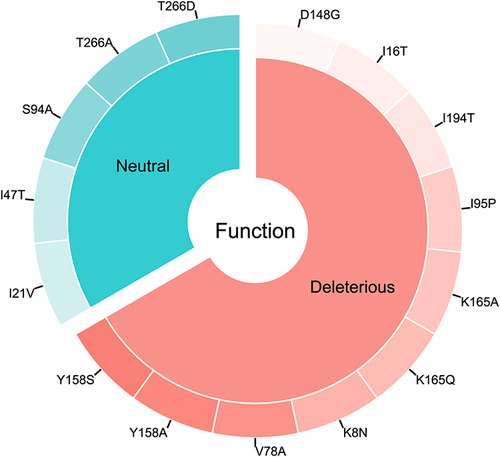
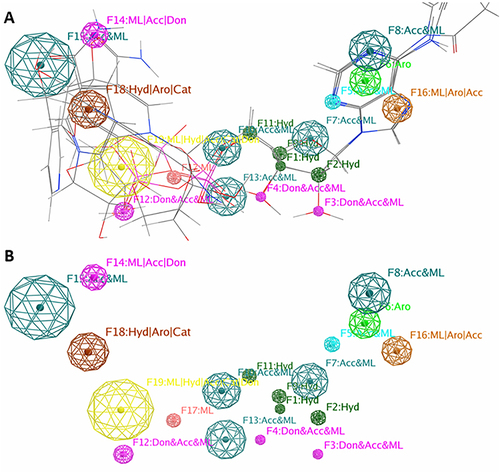
Figures & data
Figure 3 (A) The effect of the deleterious mutations on solvent accessibility surface area (SASA) between the wild-type protein (in blue) and the mutanttype protein (in orange). (B) The effect of the deleterious mutations on the flexibility (in orange) and stability (in blue) of the InhA protein.
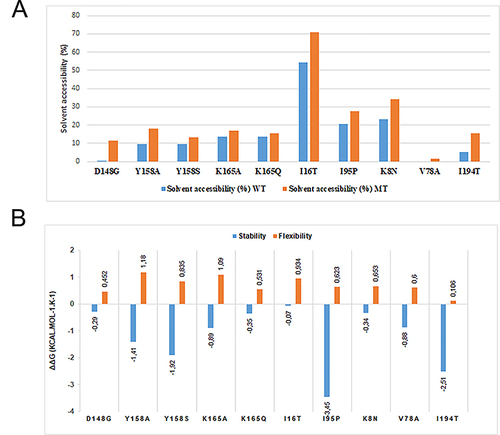
Figure 4 The three best leads with lowest affinity binding to the wild-type protein and the the mutant models.
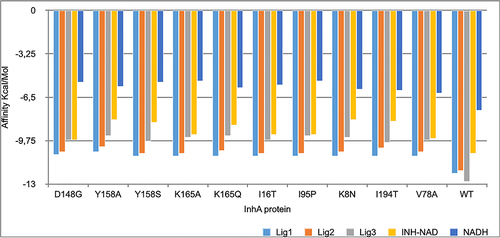
Table 1 The essential information about the reference ligand INH-NAD, natural ligand NADH and the Three best ligands including molecular formula, binding affinity score, hydrogen bond CID in PubChem, molecular weight, and toxicity test
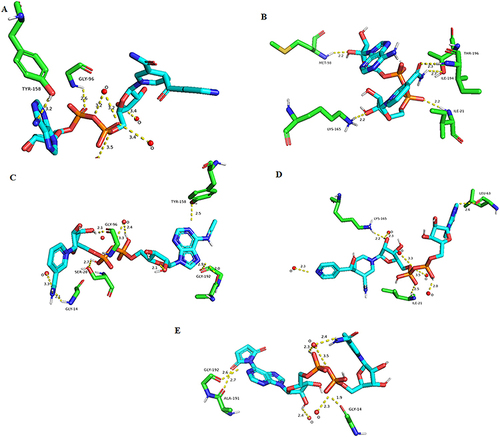
Figure 5 Visualization of 3D interactions against InhA protein. (A) Interaction between the reference ligand Inh-NAD and the inhA protein. (B) Interactions between NADH and InhA protein. (C) Interaction between ligand (CID:25176455) and inhA protein. (D) Interaction between ligand (CID:91754233) and InhA protein. (E) Interaction between ligand (CID:101256471) and InhA protein.