Figures & data
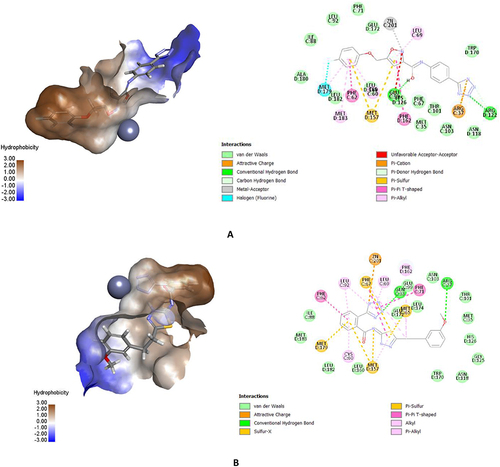
Figure 2 The active site of Glo-I enzyme; (A) Positively ionized mouth (blue), hydrophobic pocket (brown) amino acids, (B) Zinc atom (grey sphere) with major amino acids bound to it (Glu99, Gln33, his126, Glu172).
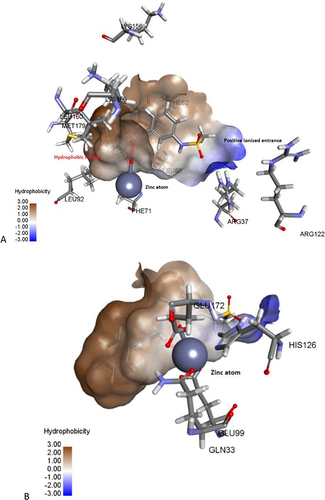
Table 1 The Values of -CDOCKER-Interaction at Two Active Sites
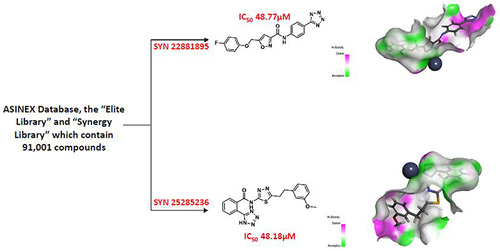
Figure 3 The chemical structures of the finally selected and purchased chemicals from the commercial database.
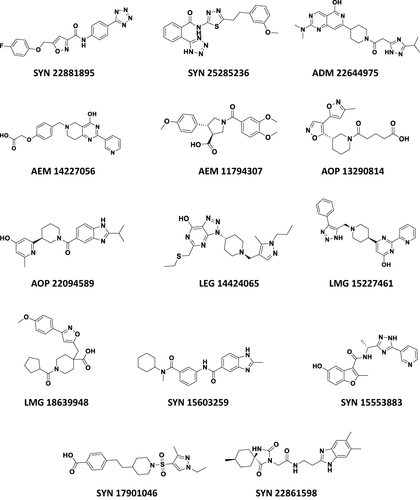
Table 2 The Values of in-situ Minimization, Binding Energy and Total Binding Energy
Table 3 The Average Percent of Inhibition and IC50 for the Tested Compounds