Figures & data
Figure 1 Overlay of H5N1 crystal structure (2hu0, yellow) and the modeled structure of H1N1 (red). The root-mean-square deviation of the two structures is 0.81 Å.
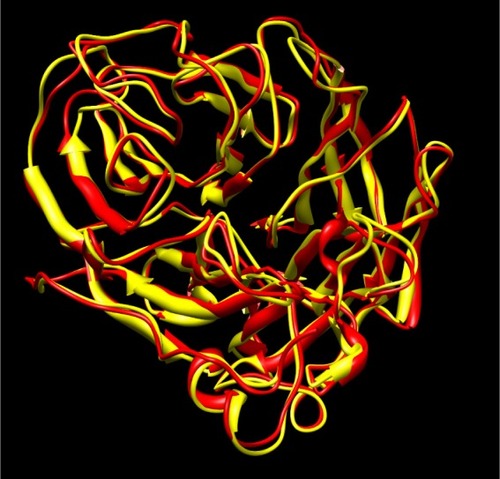
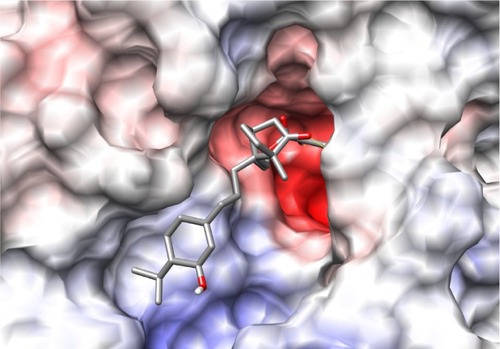
Figure 2 Picture depicting the two regions of the H1N1 neuraminidase binding site. The red region (A) is the electronegative region, where the amine-rich functional groups of ligands bind, and the large blue region (B) a large electropositive zone. The deep binding pocket (A) could host many kinds of ligands, but high affinity can be obtained only when there is a well defined functional group interaction (electrostatic and/or structural).
Table 1 Docking energies (affinity) for different chemicals with H1N1 neuraminidase
Figure 5 Chemical structure of combination ligands. Methyl salicylate-menthol-camphor (A), methyl salicylate-camphor-menthol (B), and camphor-methyl salicylate-menthol (C). The arrows show the methylene bridges artificially created to enable the possibility of docking the combination ligand with the enzyme. Note the drop in docking energy of B (−11.18 kcal/mol) when menthol and camphor position is interchanged from A (−12.68).