Figures & data
Figure 1 Structure variability within a protein family.
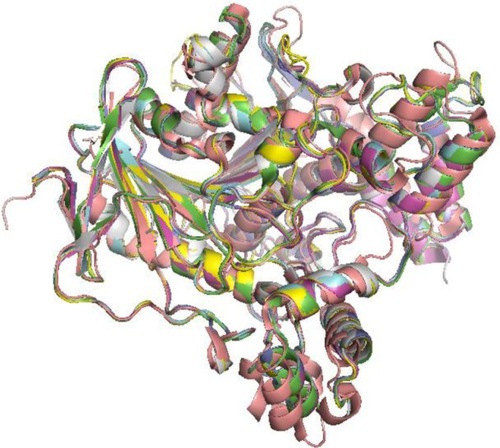
Figure 2 Experimental ensembles.
Abbreviations: NMR, nuclear magnetic resonance; PDB, protein data bank.
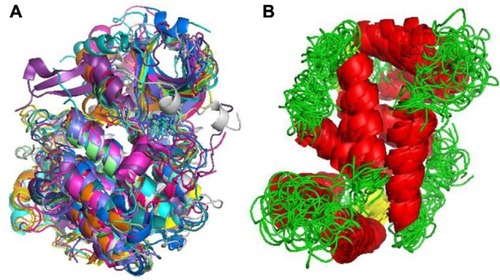
Figure 3 Molecular dynamics basic algorithm.
Abbreviations: Epot, potential energy; t, simulation time; dt, iteration time; For each spatial coordinate of the N simulated atoms (i): x, atom coordinate; F, forces component; a, acceleration; m, atom mass; v, velocity.
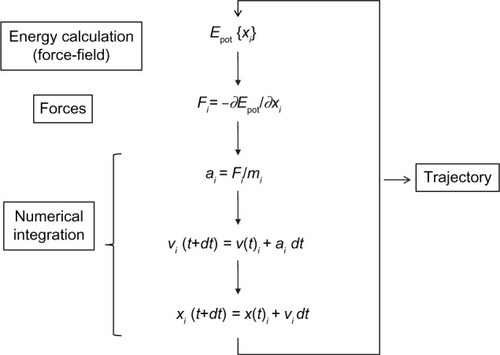
Figure 4 R-T transition on Bacillus stearothermophilus lactate dehydrogenase after 50 ns simulation in explicit solvent.
Abbreviation: R-T, relaxed and tense states (as defined by Monod’s model).
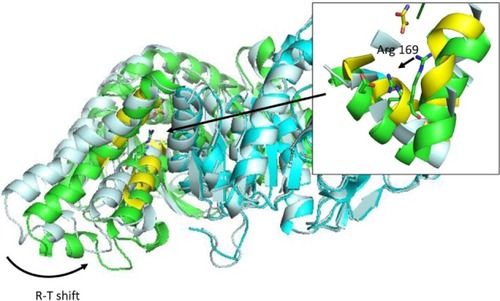
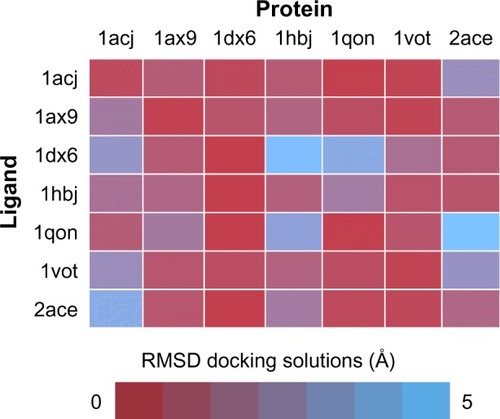
Figure 5 Cross-docking experiment with selected acetylcholinesterase structures from PDB.
Abbreviations: PDB, protein data bank; RMSD, root-mean-square deviation.