Figures & data
Table 1 Pipeline software versions used
Table 2 Population distribution in the three sets of genome assemblies
Figure 1 Normalized coverage distributions.
Abbreviations: LCL, lymphoblastoid cell line; MC1 and MC2, matched control sets 1 and 2.
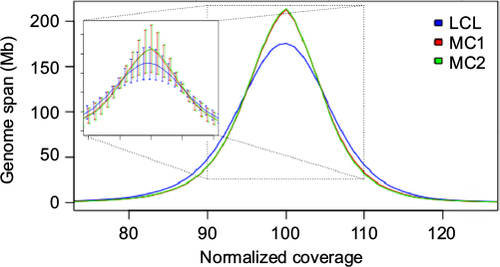
Figure 2 Software version effect.
Abbreviations: LCL, lymphoblastoid cell line; MC1 and MC2, matched control sets 1 and 2.
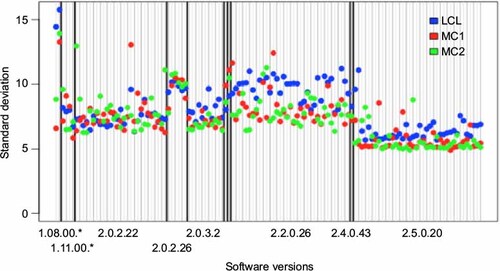
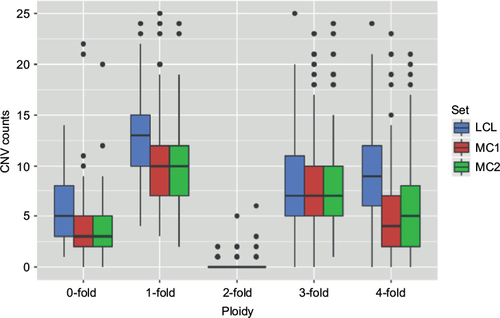
Figure 3 Chromosomal distribution of coverage deviations.
Abbreviations: LCLs, lymphoblastoid cell lines; MC1 and MC2, matched control sets 1 and 2.
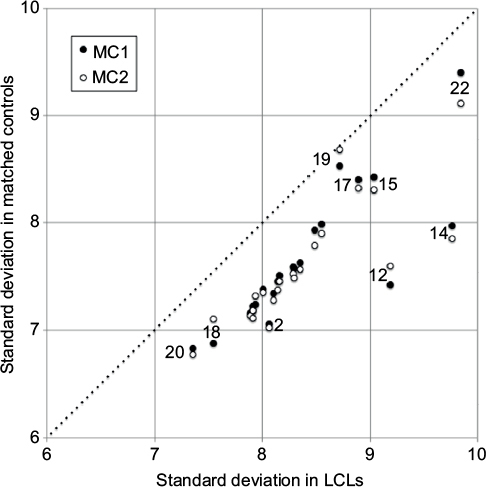
Figure 4 Genome set comparison along chromosomes.
Abbreviations: KS, Kolmogorov–Smirnov; LCLs, lymphoblastoid cell lines; MC, matched controls.
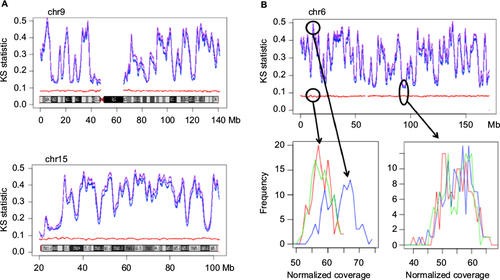
Figure 5 Correlation with various genomic parameters.
Abbreviations: KS, Kolmogorov–Smirnov.
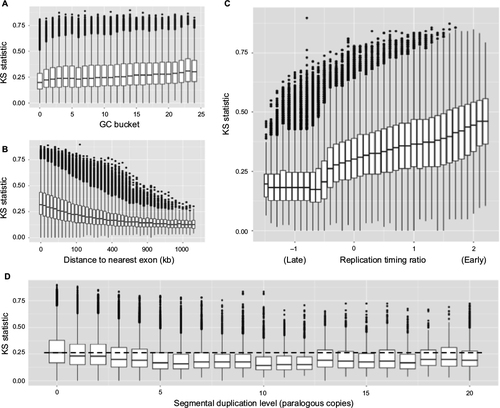
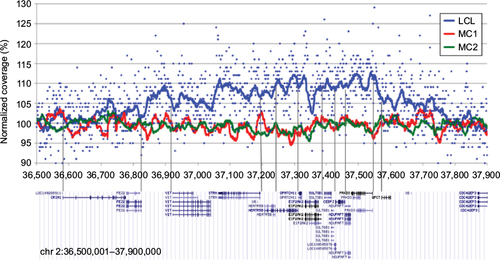
Figure 6 Example of regional coverage distortion.
Abbreviations: LCLs, lymphoblastoid cell lines; MC, matched controls.