Figures & data
Table 1 HER2 breast cancer cell-line panel
Figure 1 Heat map of maximum cell-growth inhibition induced by the individual compounds in the various cell lines.
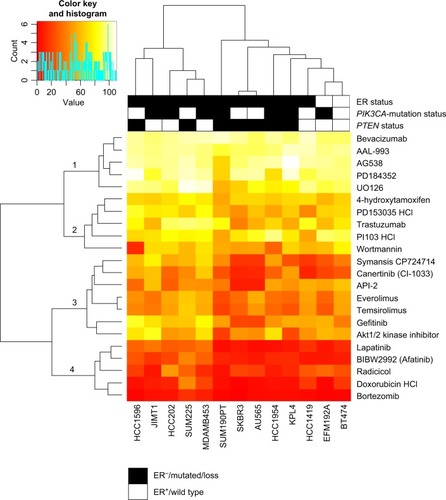
Figure 2 Cell lines divided into three groups based on response to trastuzumab and lapatinib.
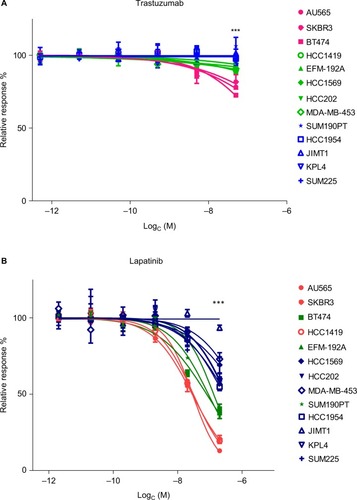
Figure 3 Box plots showing trastuzumab and lapatinib response in 13 HER2 positive cell lines relative to PIK3CA and PTEN status.
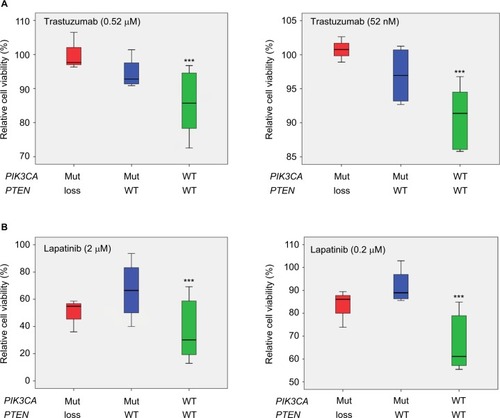
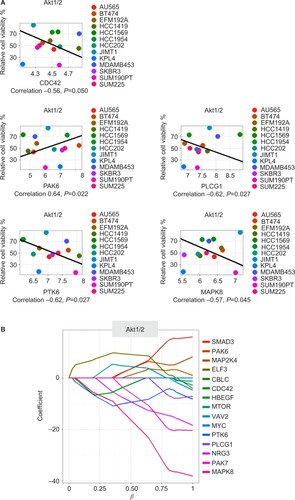
Figure 4 Akt1/2 kinase-inhibitor response.
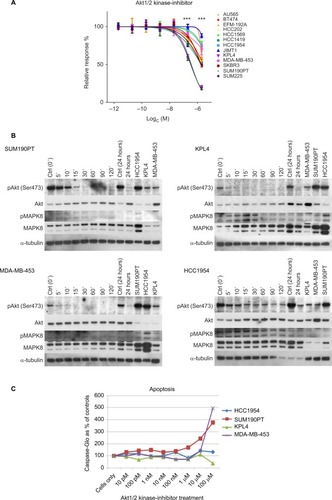
Figure 5 The five drug–gene associations found by both Spearman’s correlation and through elastic-net analysis for the Akt1/2 kinase inhibitor.