Figures & data
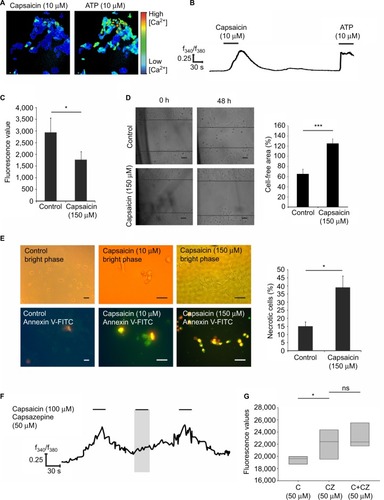
Figure 1 Heat map of TRP channel expression in 11 native human breast cancer tissues and four healthy breast tissues.
Abbreviations: CCG, Cologne Center for Genomics; FPKM, fragments per kilobase of exon per million fragments mapped; TRP, transient receptor potential.

Figure 2 Heat map of TRP channel expression in 49 breast cancer cell lines and healthy tissues.
Abbreviations: FPKM, fragments per kilobase of exon per million fragments mapped; TRP, transient receptor potential.
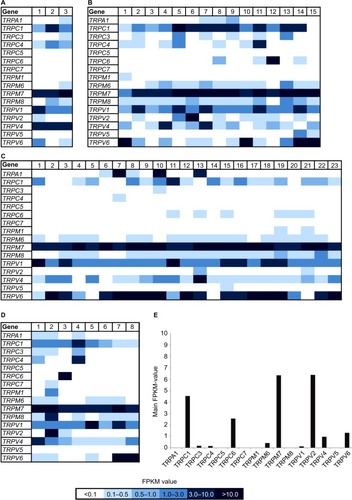
Figure 3 Expression of TRPV1 in breast cancer tissues and cell lines.
Abbreviations: BC, breast cancer; M, marker; PCR, polymerase chain reaction; RT, reverse transcriptase; TNBC, triple-negative breast cancer.
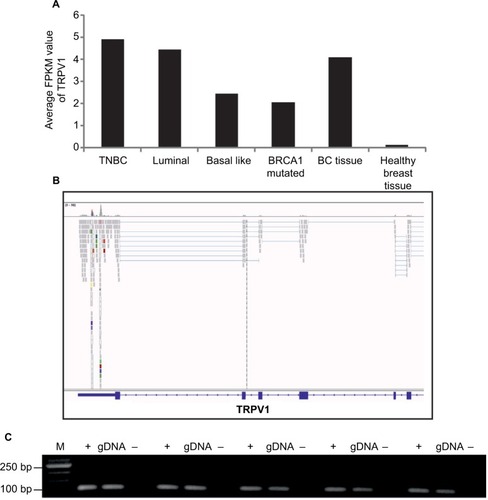
Figure 4 Expression profiles of TRP channels in the SUM149PT breast cancer cell line.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; M, marker; NGS, next-generation sequencing; PCR, polymerase chain reaction; RT, reverse transcriptase; TRP, transient receptor potential.
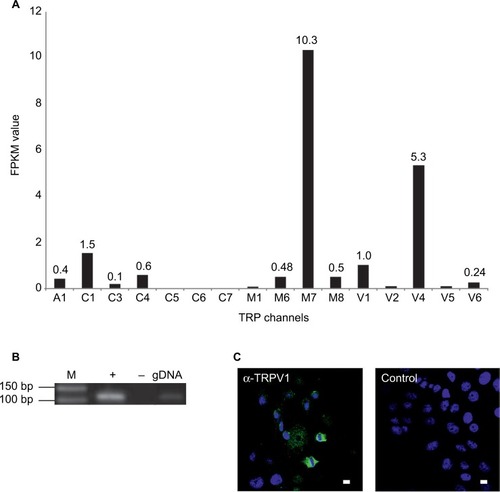
Figure 5 Functional characterization of TRPV1 activation by capsaicin in SUM149PT cells.
Abbreviations: C, capsaicin; CZ, capsazepin; FITC, fluorescein isothiocyanate; PI, propidium iodide; F340/F380, ratio of the fluorescence intensities at 340 nm and 380 nm excitation; C, capsaicin; CZ, capsazepin; ATP, adenosine triphosphate; ns, not significant.