Figures & data
Figure 1 Structure of class IA PI3K. Class IA PI3Ks are heterodimers consisting of p110 and p85 subunits. There are three p110 catalytic isoforms: p110α, p110β, and p110δ. The p110 isoforms share five distinct domains: an amino-terminal p85-binding domain (p85 BD), an RAS-binding domain (RAS BD), a putative membrane-binding domain (C2), the helical domain, and the carboxy-terminal kinase catalytic domain. There are also three p85 isoforms: p85α (and its splice variants p55α and p50α), p85β, and p55γ. They share three core domains, including a p110-binding domain called the inter-Src homology 2 (iSH2) domain, along with two SH2 domains. The two longer isoforms, p85α and p85β, have an SH3 domain and a BCR homology domain (BHD) located in their extended N-terminal regions.
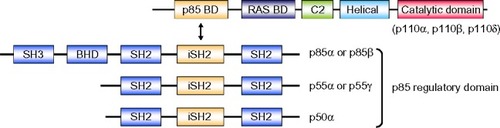
Figure 2 Class I PI3K pathway. RTK activation allows p85 to interact with RTK directly or via adaptor proteins, which recruits PI3K to the membrane. On the cell membrane, inhibitory regulation of p85 to 110 is canceled, and PI3K becomes active as a kinase. Subsequently, PI3K catalyzes the conversion of PIP2 to PIP3. PTEN catalyzes the conversion of PIP3 to PIP2. PIP3 is further recognized by AKT and PDPK1. The connection of PIP3 to PDPK1 and AKT allows the physical interaction of PDPK1 and AKT, which leads to activation of AKT by phosphorylation of the T308 residue. For maximal activation of AKT, phosphorylation of the S473 residue by mTORC2 is required. AKT phosphorylates GSK3, FOXO1, MDM2, BIM, and BAD. AKT also phosphorylates and inactivates TSC2, which subsequently allows RHEB to activate mTORC1.
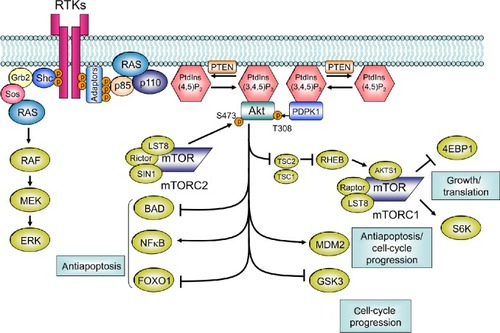
Table 1 Studies evaluating prognostic impact of PIK3CA mutations with recurrence-free survival (RFS) as end point
Table 2 Summary of predictive value of PIK3CA mutations in clinical settings