Figures & data
Figure 1 Conceptual model of using variability in genes evaluated as part of prognostic multigene expression profiles for breast cancer to test the hypothesis that heterogeneity in the biology of breast cancers at the cellular level could account for symptom variation.
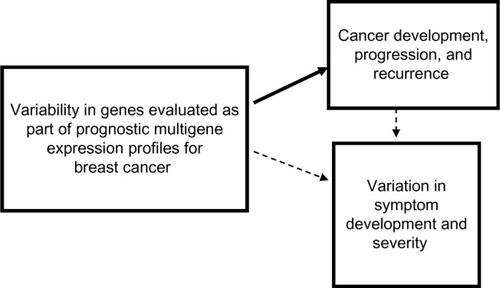
Table 1 Prognostic multigene expression profiles
Figure 2 Overlapping canonical pathways map representing shared biology among the identified candidate genes.
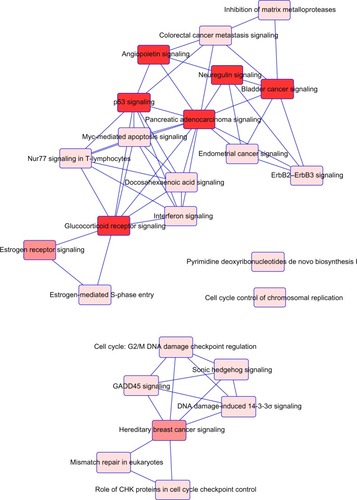
Figure 3 Gene–gene networks generated by pathway analysis.
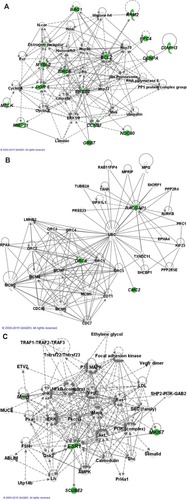
Table 2 Genes utilized in two or more prognostic multigene expression profiles as indicated by X
