Figures & data
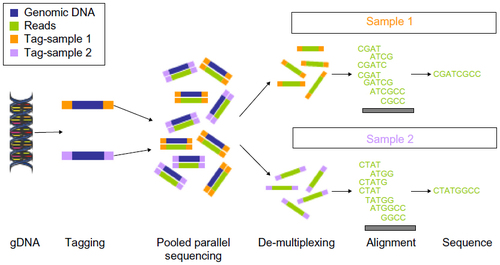
Figure 1 Translocations involving the IGH locus in mature B-cell malignancies.
Notes: (A) Summary of the different B-cell neoplasms, the normal cellular counterparts they arise from, and the main translocations involving the IGH locus they harbor (the disease-defining translocations are shown in red). (B) Translocations involving the IGH locus in mature B-cell malignancies: putative genes involved and frequencies.
Abbreviations: ABC, activated B-cell; ALL, acute lymphocytic lymphoma; B-ALL, B acute lymphoblastic leukemia; BM, bone marrow; CLL, chronic lymphocytic lymphoma; CSR, class switch recombination; DLBC, diffuse large B-cell lymphoma; FL, follicular lymphoma; GC, germinal center; Ig, immunoglobulin; IGH, immunoglobulin heavy chain; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; SLL, small lymphocytic lymphoma; SHM, somatic hypermutation; V(D)J, variable (V), diversity (D), joining (J) segments of the immunoglobulin gene; MALT, mucosa-associated lymphoid tissue.
Abbreviations: ABC, activated B-cell; ALL, acute lymphocytic lymphoma; B-ALL, B acute lymphoblastic leukemia; BM, bone marrow; CLL, chronic lymphocytic lymphoma; CSR, class switch recombination; DLBC, diffuse large B-cell lymphoma; FL, follicular lymphoma; GC, germinal center; Ig, immunoglobulin; IGH, immunoglobulin heavy chain; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; SLL, small lymphocytic lymphoma; SHM, somatic hypermutation; V(D)J, variable (V), diversity (D), joining (J) segments of the immunoglobulin gene; MALT, mucosa-associated lymphoid tissue.
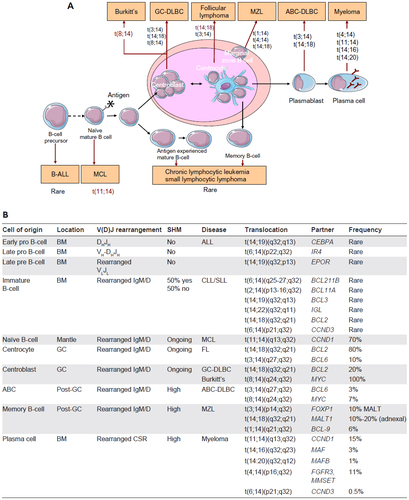
Figure 2 Heavy chain rearrangement.
Notes: In the human genome, there are approximately 50 VH, 25 DH, and six JH segments. First the DJ segments are joined, then a V segment joins to the combined DJ segments. This process is under the control of the RAG genes. Any V(D)J segment formed may be expressed with a CH region via a process of DNA rearrangement referred to as class switch recombination in the germinal center creating additional diversity where different effectors functions are associated with different C regions. V(D)JC transcript combination of a V, D, J and C (constant) region.
Abbreviation: V(D)J, variable (V), diversity (D), joining (J) segments of the immunoglobulin gene.
Abbreviation: V(D)J, variable (V), diversity (D), joining (J) segments of the immunoglobulin gene.
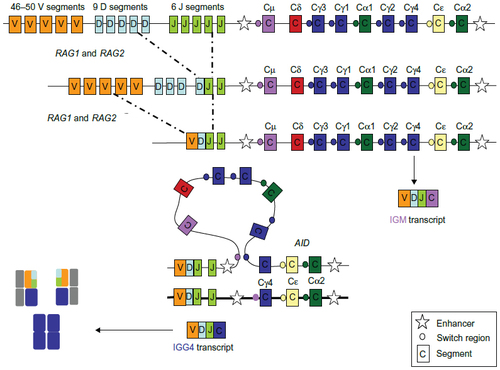
Figure 3 Overview of high throughput next-generation sequencing.
Notes: Tagging: two genomic DNA fragments (gDNA), from two distinct samples are attached to a barcode that identifies the sample from which it originates. Pooled parallel sequencing: Libraries from each sample are pooled and sequenced in parallel. Each new read contains both the DNA sequence and the barcode. Demultiplexing: Barcodes are used to distinguish reads from both samples. Alignment: Each set of reads is aligned to the reference sequence.