Figures & data
Figure 1 (A) Pedigrees and Sanger sequencing of families with EDA mutation:□, normal male; ○, normal female; ◉, female XLHED carrier; ■, male XLHED patient; the arrow shows the proband; pedigree with those affected shown in black filled symbols. (B) Sequence chromatograms (The Red arrowhead denotes that the novel missensemutationcan Resulting in the replacement of methionine at codon 373 with isoleucine.):(a) Hemizygousmutation. (b) Heterozygous mutation. (c) Normal DNA sequence. (C) Multiple sequence alignment of EDA from a variety of species. The red rectangular frames indicate the locations of M373I.
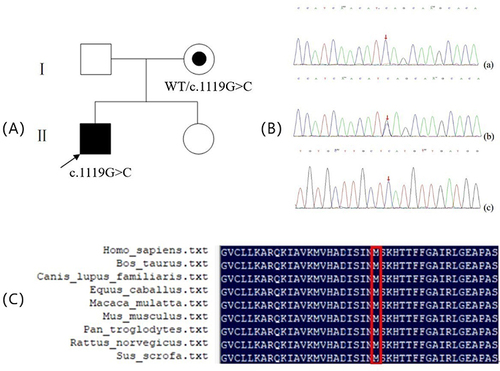
Figure 2 (A) Real-Time Quantitative PCR:Compared with wild-type EDA, the mutant EDA protein significantly inhibited the expression of EDA level.****means p <0 .0001 (Student t test). (B and C) Protein expression of mutant EDA1 in transfected cell.HEK293T cells were transfected with vectors encoding mutant or wild-type soluble FLAG-tagged EDA1 protein, cell lysates were separately analyzed by western blotting, β-tubulin was used as a loading control. The bands showed that wild-type EDA1 can produce proteins, the weaker bands of p.M373I mutations in the cell lysates showed decreased intracellular protein expression. ****means p <0 .0001 (Student t test).
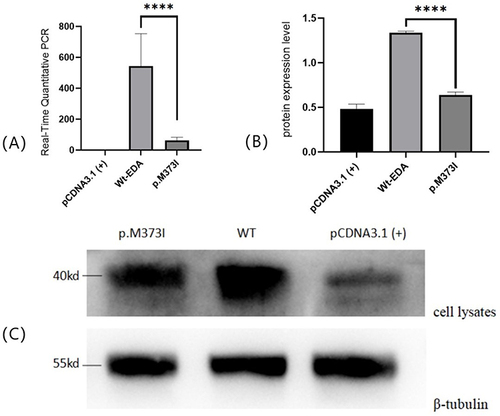
Figure 3 NF- κB transcriptional activation of mutant EDA. The mutant EDA protein attenuated transcriptionalNF-κB activation compared to wild- type EDA. **means p <0 .05 (Student t test).
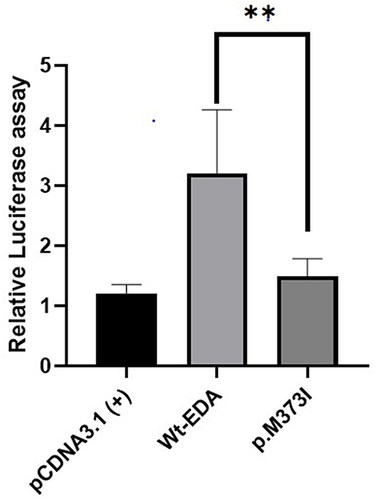
Table 1 EDA Gene Mutations Reported as of 2021 According to HGMD Database
Table 2 Summary of the Clinical Features and Genetic Data of Our Cases and Reported Cases in the Last Five Years
Data Sharing Statement
The data analyzed in this study are available from the appropriate authors upon reasonable request.
