Figures & data
Table 1 DNA repair and DNA damage response genes
Table 8 Genes associated with protection against environmental and metabolic toxicity
Figure 1 The damaging effects of dietary factors and inflammatory conditions on the colonic epithelium. Damage to DNA, the mitotic spindle, and to telomeres is mediated through the generation of ROS (reactive oxygen species) and/or RNS (reactive nitrogen species). This damage results in the activation of spindle and DNA damage checkpoints, which delay mitosis until repairs are made.
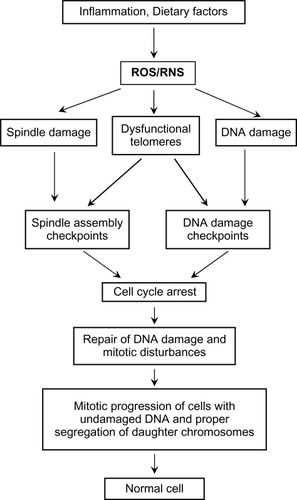
Figure 2 Excessive spindle damage, dysfunctional telomeres, or DNA damage can result in a prolonged cell cycle arrest which activates pro-cell death pathways. This activation of pro-cell death pathways leads to removal of cells with unrepaired damage to the mitotic spindle, the chromosome ends, and DNA and prevents the potential propagation of cells with many types of genomic instability.
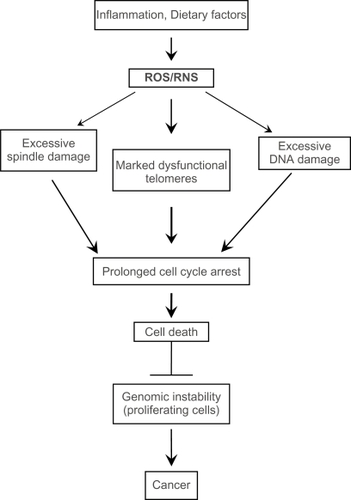
Figure 3 Examples of cellular alterations that accompany apoptosis (A), mitotic perturbation during anaphase (B), mitotic catastrophe with complete chromosome/spindle disruption (C), and abundant micronuclei formation associated with aneuploidy (D). Panels A, B, and D are examples of HCT-116 cells treated with 10 μM camptothecin. Panel C represents cells treated with 5 μM phenstatin (drug obtained through courtesy of Dr GR. Pettit, Arizona State University) (cytospin preparations of Giemsa-stained cells; ×100 oil objective lens)
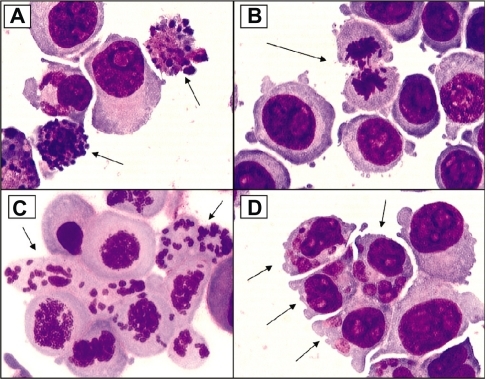
Figure 4 DNA damage causes several downstream molecular and cellular events. The DNA damage response involves several DNA repair proteins and transcription factors that allow the cell cycle to be arrested at several points to enhance genomic stability. All of the genes associated with these damage response pathways that are also found on chromosome 1p are highlighted in red, and reference to the appropriate tables (contain functions of gene products) in the text is provided below. The large number of molecular and cellular events affected by the loss of chromosome 1p is apparent.
Abbreviations: DDR, DNA damage response; ROS, reactive oxygen species; RNS, reactive nitrogen species.
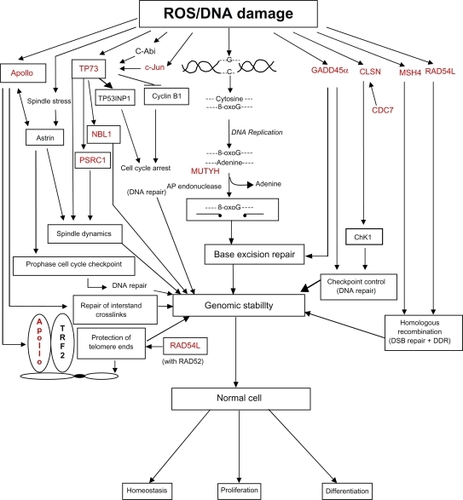
Table 2 Mitosis-related and spindle checkpoint genes
Table 6 Tumor suppressor genes
Table 7 Genes associated with antioxidant function
Figure 5 The mechanisms by which excessive activity of MUTYH and AP endonucleases can lead to cell death through the activation of PARP and the generation of toxic poly(ADP)ribose (PAR) polymers and mitochondrial DNA (mtDNA) damage (see text for detailed description).
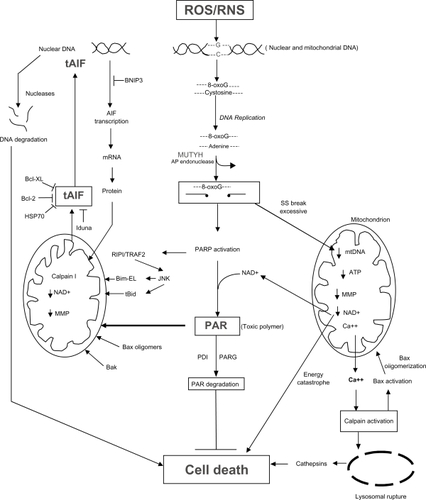
Figure 6 The possible mechanisms by which p73 transcription and activation can lead to cell death through classic apoptotic mechanisms. Definitions of proteins not included in the main text: PERP (p53 apoptosis effector related to PMP22; tetraspan membrane protein and component of intercellular desmosome junctions); p53AIPI (p53 apoptosis-inducing protein 1; promoter activated by acetylated p73); FAS (CD95) (member 6 of the TNF receptor superfamily which contains a death domain); TNFR1 (member 1A of the TNF receptor superfamily); TRAIL-R1 (member 10A of the TNF receptor superfamily); TRAIL-R2 (member 10B of the TNF receptor superfamily; death receptor 5); PIG3 (p53-induced gene 3 protein; quinone oxidoreductase involved in the generation of ROS and cell death).
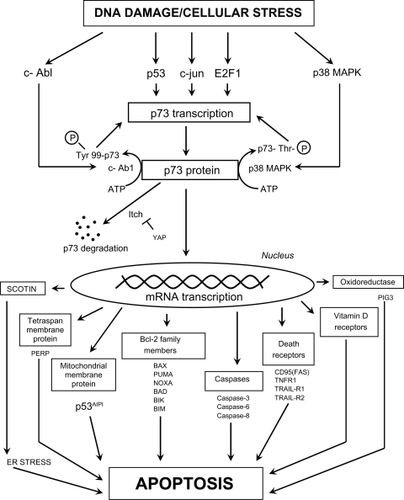
Figure 7 The different cellular fate following spindle, telomere and DNA damage during mitosis. Cells with excessive genomic damage can undergo caspase-dependent cell death (CDMCD) or caspase-independent mitotic cell death (CIMCD). DNA-damaged cells may, however, exit from mitosis by defying cell death pathways through a process referred to as mitotic slippage. These preneoplastic cells with DNA damage and chromosomal abnormalities can then be clonally expanded to produce a tumor and eventually develop into a malignancy through continued cycles of damage to the genome.
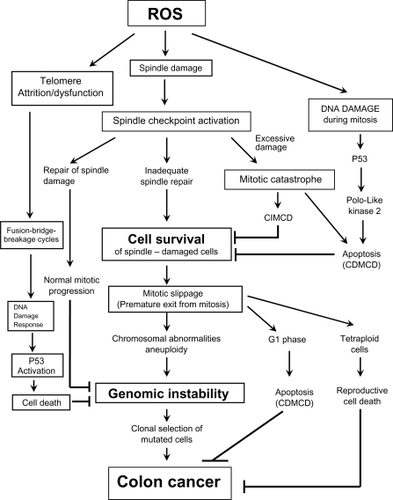
Table 3 Apoptosis-related genes
Figure 8 Transmission electron micrographs of HCT-116 cells reacted with 0.5 mM sodium deoxycholate for 2 hours. A) Normal cell (arrow 1) with prominent nucleolus and dispersed chromatin; arrow 2 points to a cell in early apoptosis, showing margination of chromatin, a nucleolus showing nucleolar segregation, and an increase in electron density compared with the normal cell; arrow 3 points to a cell in a late stage of apoptosis showing condensed chromatin, a marked increase in electron density compared with the cell above, and apoptotic body formation. B) Apoptotic cell in a late stage of apoptosis showing condensed chromatin (including crescent formation), an increase in electron density, and cytoplasmic vacuole (V) formation. (Uranyl acetate, lead citrate stains.)
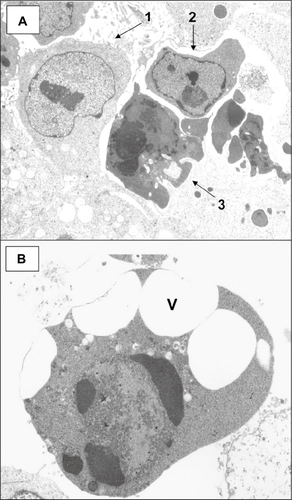
Table 4 MicroRNAs (miRNAs) and components of the miRNA processing complex
Table 5 Genes associated with the Wnt signaling pathway
Table 9 Summary of pro-cell death genes on chromosome 1p
Table 10 Preventive effects of dietary factors on processes and signaling pathways associated with genes located on chromosome 1p