Figures & data
Figure 1 Genotyping of XRCC1 arg194trp and arg399gln; XPC poly AT insertion/deletion indicated as pat; XPD arg156arg (allele C or A); GSTP1 Ile105Val and Ala114Val. (A) PCR products for XPC pat alleles: pat −/− generate the 266 bp fragment, pat +/+ generate the 344 bp fragment; heterozygous +/− were represented by a 266 bp plus a 344 bp DNA fragments. (B) The XPD arg156arg SNP is represented as nucleotide, which we identified in C/C alleles which gave 536 bp, C/A alleles with 536 bp, 363 bp and 173 bp fragments and A/A alleles with 363 bp and 173 bp fragments. (C) the XRCC1 arg194arg is indicated by a 375 bp fragment while the arg194trp is indicated by a 375 bp, 254 bp and 121 bp fragments. The XRCC1 arg399arg produced the 445 bp fragment, the arg399gln produced the 445 bp, 255 bp and 190 bp fragments, and gln399gln produced 255 bp and 190 bp fragments. (D) the SNPs in detoxifying gene GSTP1: ala114ala gave the 192 bp fragment, ala114val the 108 bp and 84 bp fragments; val114val the 192 bp, 108 bp and 84 bp fragments. (E) GSTP1 ile105ile was determined by the 216 bp fragment, the ile105val by the 121 bp and 95 bp fragments, the val105val by the 216 bp, 121 bp and 95 bp fragment.
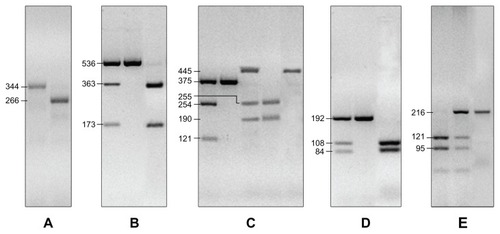
Table 1 Distributions of sex, smoking history, and polymorphisms among cases and controls
Table 2 r-value from linear correlation
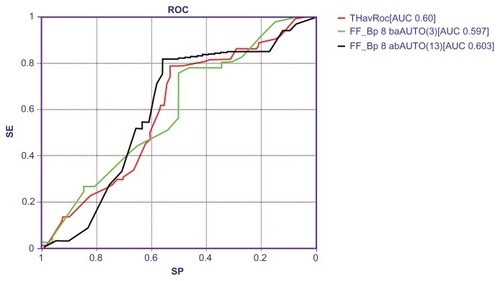
Table 3 Variables selected by the TWIST system
Table 4 Classification performances of back-propagation neural networks on final data set