Figures & data
Table 1 Summary of the GLUT family proteins and their characteristics
Figure 1 Amino acid sequences alignments of the GLUT family of proteins.
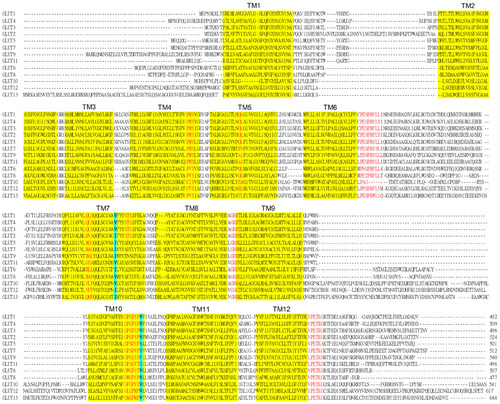
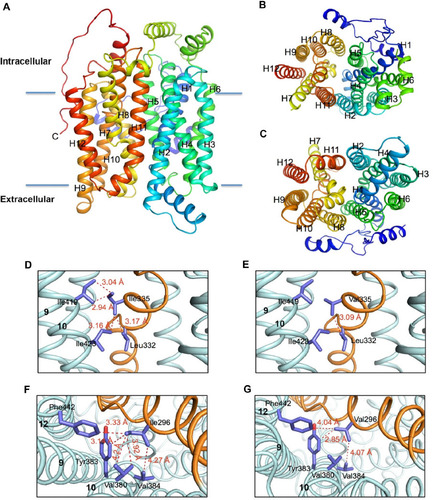
Figure 2 Molecular models of the human hGLUT9 and hGLUT5 transporters comparing possible hydrophobic interactions.