Figures & data
Table 1 Clinicopathologic variables in 83 gastric cancer patients
Figure 1 GATA3 is downregulated in gastric cancer tissues.
Abbreviation: qRT-PCR, quantitative real time polymerase chain reaction.
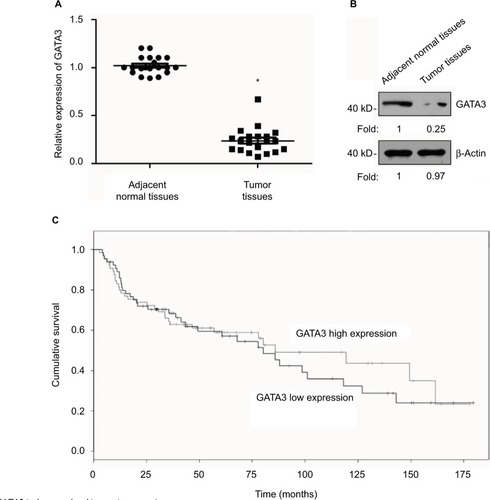
Figure 2 GATA3 is downregulated in GC cells.
Abbreviations: GC, gastric cancer; SCR, scramble siRNA; qRT-PCR, quantitative real time polymerase chain reaction.
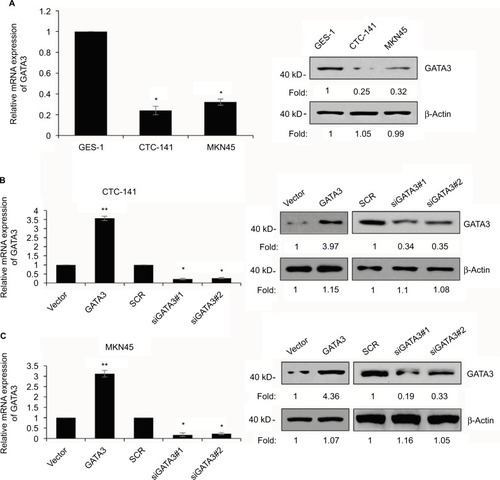
Figure 3 Restoration of GATA3 levels inhibits the proliferation of gastric carcinoma cells.
Abbreviations: CCK-8, Cell Counting Kit-8; SCR, scramble siRNA.
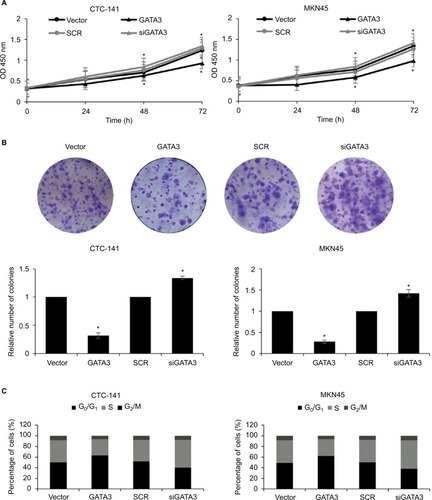
Figure 4 Restoration of GATA3 levels inhibits the migratory and invasive behavior of gastric cancer cells.
Abbreviation: SCR, scramble siRNA.
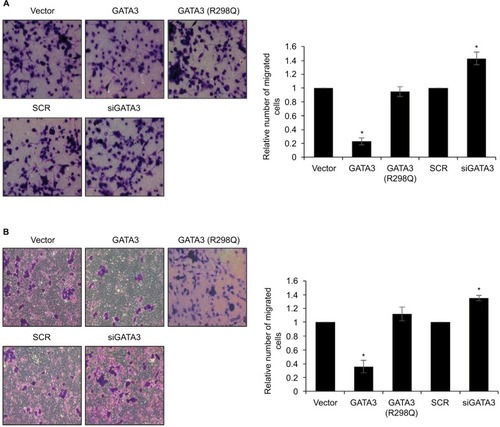
Figure 5 GATA3 inhibits epithelial–mesenchymal transition (EMT) in gastric cancer cells.
Abbreviations: SCR, scramble siRNA; qRT-PCR, quantitative real time polymerase chain reaction.
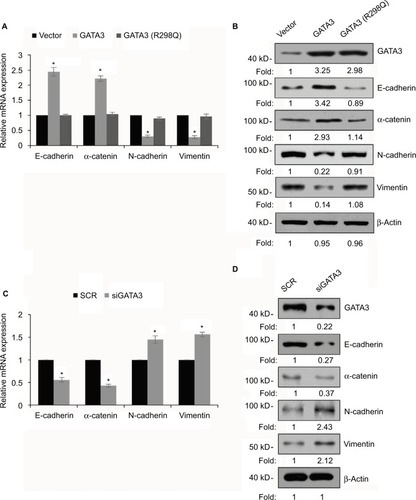
Figure 6 GATA3 transcriptionally regulates ZEB1 in G cells.
Abbreviations: qRT-PCR, quantitative real time polymerase chain reaction; ChIP, chromatin immunoprecipitation; qChIP, quantitative chromatin immunoprecipitation.
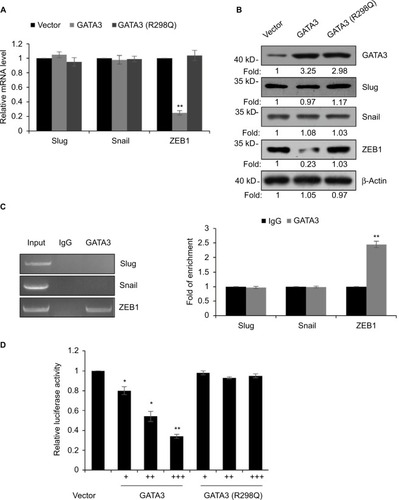
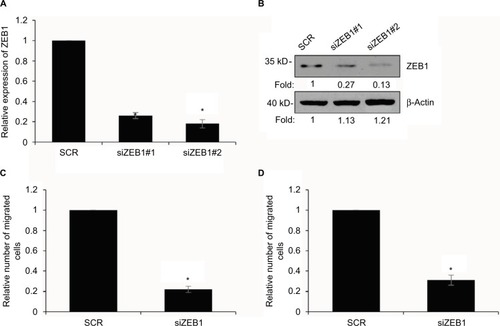
Figure 7 Overexpression of ZEB1 suppresses the migratory and invasive behavior of gastric cancer cells.
Abbreviations: SCR, scramble siRNA; qRT-PCR, quantitative real time polymerase chain reaction.