Figures & data
Table 1 Correlations of sirtuins and clinicopathological parameters in patients with NSCLC
Table 2 Cox proportional regression analysis to investigate the correlations of sirtuins and overall survival in patients with NSCLC
Table 3 Cox proportional regression analysis to investigate the correlations of sirtuins and recurrence-free survival in patients with NSCLC
Figure 1 Overall survival in TCGA NSCLC patients with high and low sirtuin expression (SIRT1–7) were compared with Kaplan–Meier plot curves.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7.
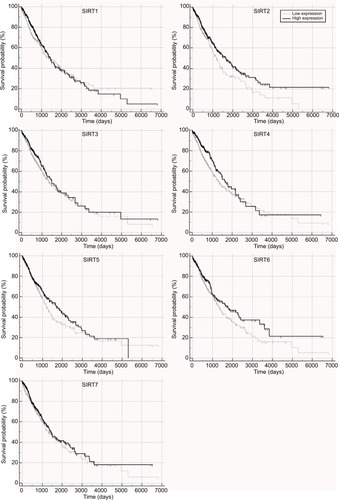
Figure 2 Recurrence-free survival in TCGA NSCLC patients stratified by high and low sirtuin expression (SIRT1–7) or a combination of SIRT1 and SIRT2 were compared with Kaplan–Meier plot curves. (A) SIRT1, (B) SIRT2, (C) SIRT3, (D) SIRT4, (E) SIRT5, (F) SIRT6, (G) SIRT7, and (H) Combination of SIRT1 and SIRT2.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; +, high expression; –, low expression.
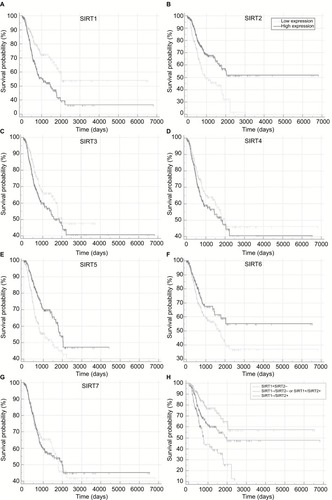
Table 4 Cox proportional regression analysis to investigate the correlations of the combination of SIRT1 and SIRT2 with recurrence-free survival in patients with NSCLC
Figure 3 Forest plot for the correlations of sirtuins (SIRT1–7) and recurrence-free survival in patients with NSCLC by pooling the results of the datasets from TCGA and Kaplan–Meier plotter databases.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7.
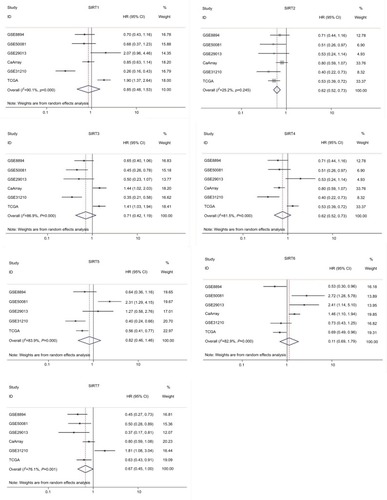
Table 5 Associations of high sirtuins expressions with overall survival in NSCLC in previous reports
Figure 4 Biological processes regulated by SIRT1, SIRT2, and SIRT7 in TCGA NSCLC patients: (A) SIRT1, (B) SIRT2, and (C). SIRT7.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT7, sirtuin 7.
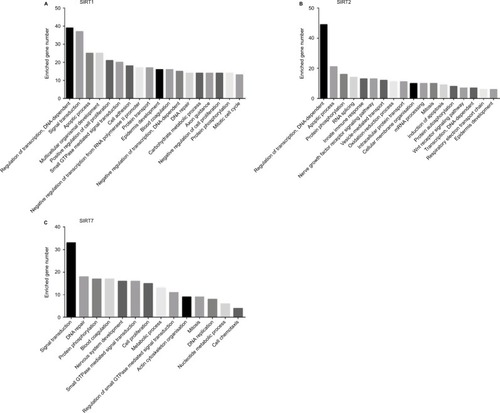
Figure 5 Expression correlations of SIRT1, SIRT2, and SIRT7 with BAD, BAX, BID, BCL2, and ZEB1 in TCGA NSCLC patients. (A–C). Correlations of SIRT1 with BAD (A), BAX (B), and BID (C). (D) Correlation of SIRT2 with BAD. (E–H) Correlations of SIRT7 with BAD (E), BAX (F), BCL2 (G), and ZEB1 (H).
Abbreviations: SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT7, sirtuin 7; BAX, Bcl-2 associated X; BAD, Bcl-2 associated agonist of cell death; BID, BH3 interacting domain death agonist; ZEB1, zinc finger E-box binding homeobox 1; TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer.
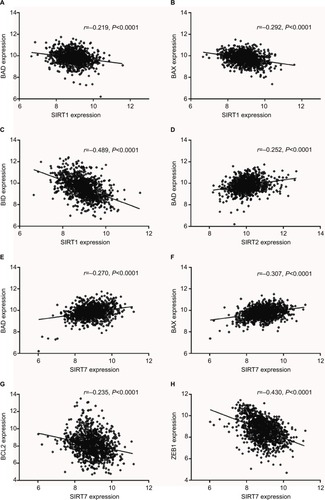
Figure 6 Concordance of SIRT2 mRNA and protein expressions in NSCLC. (A). SIRT2 immunohistochemical staining results in normal lung tissues (n=3) and lung cancer tissues (n=12). The images were obtained from the Human Protein Atlas.Citation43 (B). SIRT2 mRNA expression in normal lung tissues (n=110) and lung cancer tissues (n=1017) in the TCGA NSCLC cohort. (C–E). SIRT2 mRNA expression detected by qRT-PCR (C) and protein expression detected by Western blotting (D) in seven lung cancers and paired adjacent normal tissues (n=7); consistency of SIRT2 mRNA and protein expression were explored by Pearson’s correlation analysis (E). These were re-analyses results of Li et al’s report.Citation44 (F). Associations of SIRT2 mRNA with protein expressions in lung cancers. SIRT2 protein expression and mRNA expression data were extracted from proteomics and gene microarray profiles, respectively, which was reported by Stewart et al.Citation46
Abbreviations: SIRT2, sirtuin 2; N, normal; T, tumor; qRT-PCR, quantitative real-time polymerase chain reaction.
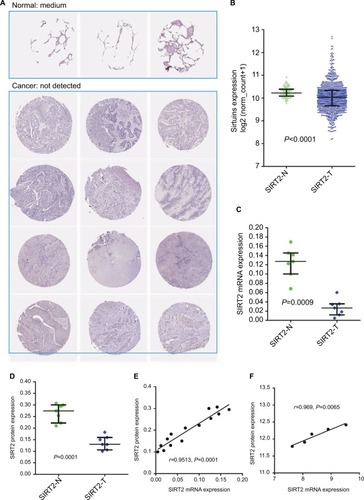
Table S1 Underlying mechanism of sirtuins in NSCLC in the previous reports
